8R45
 
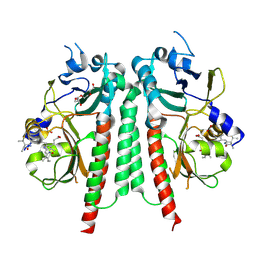 | | Phytochromobilin-adducted PAS-GAF bidomain of Glycine max phytochrome A | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A-2, TETRAETHYLENE GLYCOL | | Authors: | Guan, K, Chen, P, Nagano, S, Hughes, J. | | Deposit date: | 2023-11-13 | | Release date: | 2025-01-08 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pr and Pfr structures of plant phytochrome A.
Nat Commun, 16, 2025
|
|
7T4E
 
 | |
7T4D
 
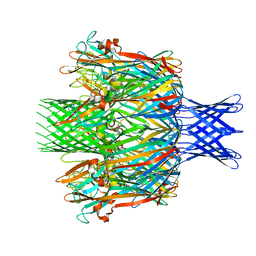
 | | Pore structure of pore-forming toxin Epx4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Epx4 | | Authors: | Xiong, X.Z, Dong, M, Yang, P, Abraham, J. | | Deposit date: | 2021-12-09 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Emerging enterococcus pore-forming toxins with MHC/HLA-I as receptors.
Cell, 185, 2022
|
|
7U1Z
 
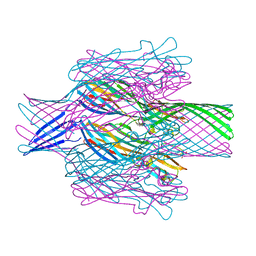
 | | Crystal structure of the DRBD and CROPs of TcdA | | Descriptor: | SULFATE ION, Toxin A | | Authors: | Baohua, C, Peng, C, Kay, P, Rongsheng, J. | | Deposit date: | 2022-02-22 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure and conformational dynamics of Clostridioides difficile toxin A.
Life Sci Alliance, 5, 2022
|
|
7XS8
 
 | |
7XSA
 
 | |
7XSC
 
 | |
7XSB
 
 | |
4JGV
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with THPN | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Li, F, Li, A, Tian, X, Wan, W, Wan, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-03-04 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4AY9
 
 | | Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLLICLE-STIMULATING HORMONE RECEPTOR, FOLLITROPIN SUBUNIT BETA, ... | | Authors: | Jiang, X, Liu, H, Chen, X, He, X. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Follicle-Stimulating Hormone in Complex with the Entire Ectodomain of its Receptor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
6Z14
 
 | | Structure of Bifidobacterium bifidum GH20 beta-N-beta-N-acetylhexosaminidase E553Q variant in complex with 4MU-6SGlcNAc-derived oxazoline | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-N-acetylhexosaminidase, NITRATE ION, ... | | Authors: | He, Y, Jin, Y, Rizkallah, P, Chen, P. | | Deposit date: | 2020-05-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the GH20 beta-N-beta-N-acetylhexosaminidase from Bifidobacterium bifidum
To Be Published
|
|
4KZM
 
 | | Crystal Structure of TR3 LBD S553A Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZJ
 
 | | Crystal Structure of TR3 LBD L449W Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZI
 
 | | Crystal Structure of TR3 LBD in complex with DPDO | | Descriptor: | 1-(3,5-dimethoxyphenyl)decan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
7FC5
 
 | | Crystal structure of SARS-CoV-2 RBD and horse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1 | | Authors: | Wang, X.Q, Lan, J, Ge, J.W. | | Deposit date: | 2021-07-13 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7FC6
 
 | | Crystal structure of SARS-CoV RBD and horse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, X.Q, Lan, J, Ge, J.W. | | Deposit date: | 2021-07-13 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
8HIH
 
 | | Cryo-EM structure of Mycobacterium tuberculosis transcription initiation complex with transcription factor GlnR | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Shi, J, Xu, J.C. | | Deposit date: | 2022-11-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the transcription activation mechanism of the global regulator GlnR from actinobacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8I7S
 
 | | The crystal structure of human abl1 kinase domain in complex with ABL1-B1 | | Descriptor: | 5-[3-(2-methoxy-5-oxidanyl-phenyl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N,N-dimethyl-pyridine-3-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z. | | Deposit date: | 2023-02-02 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94635463 Å) | | Cite: | Rationally designed BCR-ABL kinase inhibitors for improved leukemia treatment via covalent and pro-/dual-drug targeting strategies.
J Adv Res, 2024
|
|
8I7Z
 
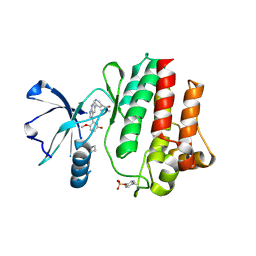 | | The crystal structure of human abl1 kinase domain in complex with ABL1-B5 | | Descriptor: | 5-[3-(2-methoxy-5-oxidanyl-phenyl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N,N-dimethyl-pyridine-3-carboxamide, BETA-ALANINE, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z. | | Deposit date: | 2023-02-02 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25495815 Å) | | Cite: | Rationally designed BCR-ABL kinase inhibitors for improved leukemia treatment via covalent and pro-/dual-drug targeting strategies.
J Adv Res, 2024
|
|
8I7T
 
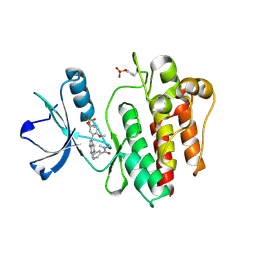 | | The crystal structure of human abl1 kinase domain in complex with ABL1-B4 | | Descriptor: | Tyrosine-protein kinase ABL1, [3-[5-[5-(dimethylcarbamoyl)pyridin-3-yl]-1H-pyrrolo[2,3-b]pyridin-3-yl]-4-methoxy-phenyl] ethanesulfonate | | Authors: | Zhu, C, Zhang, Z. | | Deposit date: | 2023-02-02 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80004954 Å) | | Cite: | Rationally designed BCR-ABL kinase inhibitors for improved leukemia treatment via covalent and pro-/dual-drug targeting strategies.
J Adv Res, 2024
|
|
7CHO
 
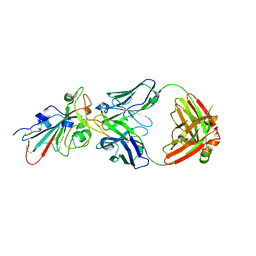 | | Crystal structure of SARS-CoV-2 antibody P5A-1D2 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-1D2 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHS
 
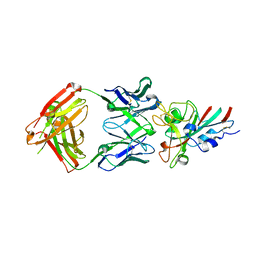 | | Crystal structure of SARS-CoV-2 antibody P22A-1D1 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P22A-1D1 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHP
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-3C8 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-3C8 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
