5ZE3
 
 | |
5ZF3
 
 | | Crystal Structures of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION, ... | | Authors: | Zhang, X, Wan, Q, Li, Z. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures of Endo-beta-1,4-xylanase II Complexed with Xylotriose
To be published
|
|
5Z93
 
 | | Crystal Structure of SIRT3 in complex with H3K9bhb peptide | | Descriptor: | Gene for histone H3 (germline gene), NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ... | | Authors: | Zhang, X, Li, H. | | Deposit date: | 2018-02-02 | | Release date: | 2019-02-06 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Crystal Structure of SIRT3 in complex with H3K9bhb peptide
To Be Published
|
|
5ZH0
 
 | | Crystal Structures of Endo-beta-1,4-xylanase II | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | Authors: | Zhang, X, Wan, Q, Li, Z. | | Deposit date: | 2018-03-10 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal Structures of Endo-beta-1,4-xylanase II
To be published
|
|
5Z94
 
 | |
5ZH9
 
 | |
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
5T2A
 
 | | CryoEM structure of the Leishmania donovani 80S ribosome at 2.9 Angstrom resolution | | Descriptor: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
5T2C
 
 | | CryoEM structure of the human ribosome at 3.6 Angstrom resolution | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
9ITO
 
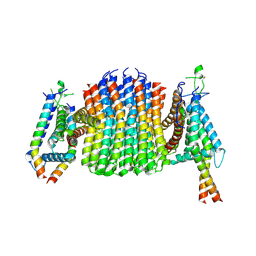 | | Chloroflexus aurantiacus ATP synthase, state 2, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITW
 
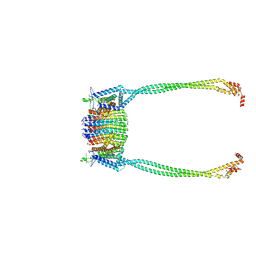 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 1, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITN
 
 | | Chloroflexus aurantiacus ATP synthase, state 1, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITX
 
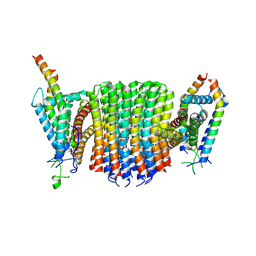 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 2, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITU
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITT
 
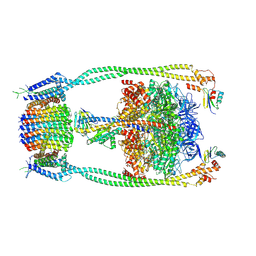 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITS
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITK
 
 | | Chloroflexus aurantiacus ATP synthase, state 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITR
 
 | | Chloroflexus aurantiacus ATP synthase, state 3, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9IU0
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 3, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (5.18 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITL
 
 | | Chloroflexus aurantiacus ATP synthase, state 3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITQ
 
 | | Chloroflexus aurantiacus ATP synthase, state 3, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITZ
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 3, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITJ
 
 | | Chloroflexus aurantiacus ATP synthase, state 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITP
 
 | | Chloroflexus aurantiacus ATP synthase, state 2, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITY
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 2, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.95 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
