1TC3
 
 | | TRANSPOSASE TC3A1-65 FROM CAENORHABDITIS ELEGANS | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*GP*TP*CP*CP*TP*AP*TP*AP*GP*A P*AP*CP*TP*T)-3'), DNA (5'-D(*AP*GP*TP*TP*CP*TP*AP*TP*AP*GP*GP*AP*CP*CP*CP*CP*C P*CP*CP*T)-3'), PROTEIN (TC3 TRANSPOSASE) | | Authors: | Van Pouderoyen, G, Ketting, R.F, Perrakis, A, Plasterk, R.H.A, Sixma, T.K. | | Deposit date: | 1997-07-07 | | Release date: | 1997-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the specific DNA-binding domain of Tc3 transposase of C.elegans in complex with transposon DNA.
EMBO J., 16, 1997
|
|
1EPU
 
 | | X-RAY crystal structure of neuronal SEC1 from squid | | Descriptor: | S-SEC1 | | Authors: | Bracher, A, Perrakis, A, Dresbach, T, Betz, H, Weissenhorn, W. | | Deposit date: | 2000-03-29 | | Release date: | 2000-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray crystal structure of neuronal Sec1 from squid sheds new light on the role of this protein in exocytosis.
Structure Fold.Des., 8, 2000
|
|
2X4N
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to residual fragments of a photocleavable peptide that is cleaved upon UV-light treatment | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2XRG
 
 | | Crystal structure of Autotaxin (ENPP2) in complex with the HA155 boronic acid inhibitor | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Hausmann, J, Albers, H.M.H.G, Perrakis, A. | | Deposit date: | 2010-09-14 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Substrate Discrimination and Integrin Binding by Autotaxin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2X70
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a photocleavable peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
1DWK
 
 | | STRUCTURE OF CYANASE WITH THE DI-ANION OXALATE BOUND AT THE ENZYME ACTIVE SITE | | Descriptor: | CYANATE HYDRATASE, OXALATE ION, SULFATE ION | | Authors: | Walsh, M.A, Otwinowski, Z, Perrakis, A, Anderson, P.M, Joachimiak, A. | | Deposit date: | 1999-12-07 | | Release date: | 2000-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Cyanase Reveals that a Novel Dimeric and Decameric Arrangement of Subunits is Required for Formation of the Enzyme Active Site.
Structure, 8, 2000
|
|
2X4U
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 Peptide RT468-476 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2XSE
 
 | | The structural basis for recognition of J-base containing DNA by a novel DNA-binding domain in JBP1 | | Descriptor: | GLYCEROL, NITRATE ION, THYMINE DIOXYGENASE JBP1 | | Authors: | Heidebrecht, T, Christodoulou, E, Chalmers, M.J, Jan, S, ter Riete, B, Grover, R.K, Joosten, R.P, Littler, D, vanLuenen, H, Griffin, P.R, Wentworth, P, Borst, P, Perrakis, A. | | Deposit date: | 2010-09-28 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Recognition of Base J Containing DNA by a Novel DNA Binding Domain in Jbp1.
Nucleic Acids Res., 39, 2011
|
|
2X4R
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to Cytomegalovirus (CMV) pp65 epitope | | Descriptor: | 65 KDA PHOSPHOPROTEIN, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2X4S
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a peptide representing the epitope of the H5N1 (Avian Flu) Nucleoprotein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals
J.Am.Chem.Soc., 131, 2009
|
|
2X4O
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 envelope peptide env120-128 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, ENVELOPE GLYCOPROTEIN GP160, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
1DW9
 
 | | Structure of cyanase reveals that a novel dimeric and decameric arrangement of subunits is required for formation of the enzyme active site | | Descriptor: | CHLORIDE ION, CYANATE LYASE, SULFATE ION | | Authors: | Walsh, M.A, Otwinowski, Z, Perrakis, A, Anderson, P.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 1999-12-03 | | Release date: | 2000-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Cyanase Reveals that a Novel Dimeric and Decameric Arrangement of Subunits is Required for Formation of the Enzyme Active Site
Structure, 8, 2000
|
|
1DCS
 
 | | DEACETOXYCEPHALOSPORIN C SYNTHASE FROM S. CLAVULIGERUS | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE, SULFATE ION | | Authors: | Valegard, K, Terwisscha Van Scheltinga, A.C, Lloyd, M.D, Hara, T, Ramaswamy, S, Perrakis, A, Thompson, A, Lee, H.J, Baldwin, J.E, Schofield, C.J, Hajdu, J, Andersson, I. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a cephalosporin synthase.
Nature, 394, 1998
|
|
1E3M
 
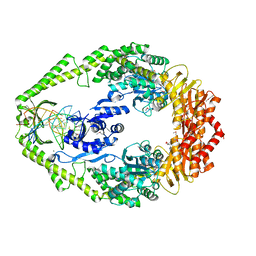 | | The crystal structure of E. coli MutS binding to DNA with a G:T mismatch | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP* GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP* TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lamers, M.H, Perrakis, A, Enzlin, J.H, Winterwerp, H.H.K, De Wind, N, Sixma, T.K. | | Deposit date: | 2000-06-19 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of DNA Mismatch Repair Protein Muts Binding to a G X T Mismatch
Nature, 407, 2000
|
|
6QC0
 
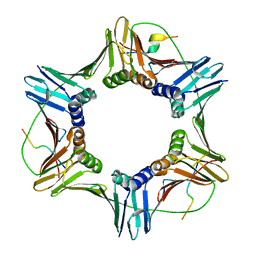 | | PCNA complex with Cdt2 C-terminal PIP-box peptide | | Descriptor: | Denticleless protein homolog, Proliferating cell nuclear antigen | | Authors: | Perrakis, A.P, von Castelmur, E. | | Deposit date: | 2018-12-25 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Direct binding of Cdt2 to PCNA is important for targeting the CRL4Cdt2E3 ligase activity to Cdt1.
Life Sci Alliance, 1, 2018
|
|
1K9T
 
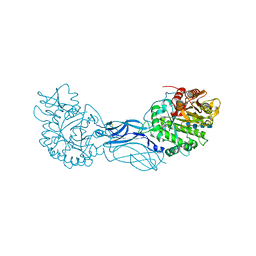 | | Chitinase a complexed with tetra-N-acetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Prag, G, Tucker, P.A, Oppenheim, A.B. | | Deposit date: | 2001-10-30 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex Structures of Chitinase A Mutant with Oligonag Provide Insight Into the Enzymatic Mechanism
To be Published
|
|
7M6B
 
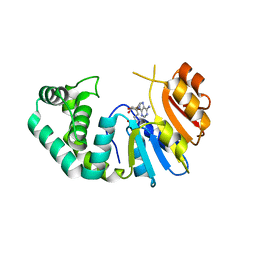 | | The Crystal Structure of Mcbe1 | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2021-03-25 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Target highlights in CASP14: Analysis of models by structure providers.
Proteins, 89, 2021
|
|
7OC9
 
 | |
8PH4
 
 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2023-06-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
8R88
 
 | |
2IUO
 
 | | Site Directed Mutagenesis of Key Residues Involved in the Catalytic Mechanism of Cyanase | | Descriptor: | AZIDE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Guilloton, M, Walsh, M.A, Joachimiak, A, Anderson, M.P. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Twin Set of Low Pka Arginines Ensures the Concerted Acid Base Catalytic Mechanism of Cyanase
To be Published
|
|
2IV1
 
 | |
1VAL
 
 | | CONCANAVALIN A COMPLEX WITH 4'-NITROPHENYL-ALPHA-D-GLUCOPYRANOSIDE | | Descriptor: | 4-nitrophenyl alpha-D-glucopyranoside, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Kanellopoulos, P.N, Tucker, P.A, Hamodrakas, S.J. | | Deposit date: | 1996-01-08 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the complexes of concanavalin A with 4'-nitrophenyl-alpha-D-mannopyranoside and 4'-nitrophenyl-alpha-D-glucopyranoside.
J.Struct.Biol., 116, 1996
|
|
1VAM
 
 | | CONCANAVALIN A COMPLEX WITH 4'-NITROPHENYL-ALPHA-D-MANNOPYRANOSIDE | | Descriptor: | 4-nitrophenyl alpha-D-mannopyranoside, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Kanellopoulos, P.N, Tucker, P.A, Hamodrakas, S.J. | | Deposit date: | 1996-01-08 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of the complexes of concanavalin A with 4'-nitrophenyl-alpha-D-mannopyranoside and 4'-nitrophenyl-alpha-D-glucopyranoside.
J.Struct.Biol., 116, 1996
|
|
9F5W
 
 | | Human condensin II - M18BP1 complex | | Descriptor: | Condensin-2 complex subunit D3, Condensin-2 complex subunit G2, Condensin-2 complex subunit H2, ... | | Authors: | Borsellini, A, Vannini, A. | | Deposit date: | 2024-04-30 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Condensin II activation by M18BP1
To Be Published
|
|
