8K9B
 
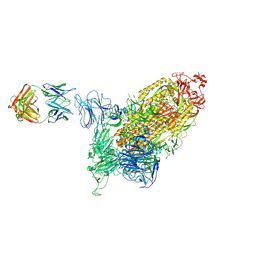 | | SARS-CoV-2 spike protein in complex with one S2H5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of S2H5 Fab, Light chain of S2H5 Fab, ... | | Authors: | Hong, Q, Cong, Y. | | Deposit date: | 2023-07-31 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis of SARS-CoV-2 immune evasion of NTD neutralizing antibodies.
Dian Zi Xian Wei Xue Bao, 2024
|
|
8K9M
 
 | |
8K9J
 
 | |
8YTC
 
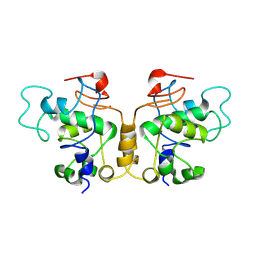 | | PML-RBCC dimer | | Descriptor: | Protein PML | | Authors: | Tan, Y, Li, J, Zhang, S, Zhang, Y, Cong, Y, Meng, G. | | Deposit date: | 2024-03-25 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of PML RBCC dimer reveals CC-mediated octopus-like nuclear body assembly mechanism.
Cell Discov, 10, 2024
|
|
7DK3
 
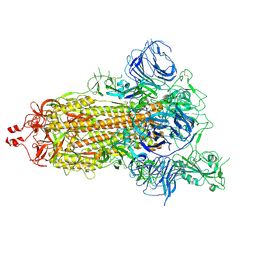 | | SARS-CoV-2 S trimer, S-open | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7YS6
 
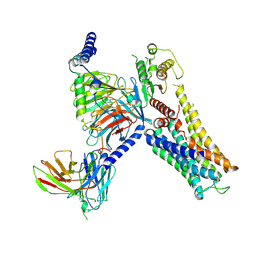 | | Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex | | Descriptor: | 5-hydroxytryptamine receptor 6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, Q.Y, Wang, Y.F, He, L, Wang, S, Cong, Y. | | Deposit date: | 2022-08-11 | | Release date: | 2023-03-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into constitutive activity of 5-HT 6 receptor.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7FIF
 
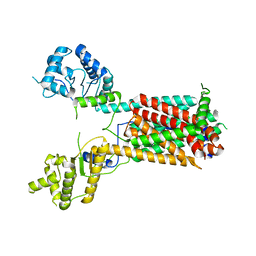 | | Cryo-EM structure of the hedgehog release protein Disp from water bear (Hypsibius dujardini) | | Descriptor: | Protein dispatched-like protein 1 | | Authors: | Luo, Y, Wan, G, Wang, Q, Zhao, Y, Cong, Y, Li, D. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Architecture of Dispatched, a Transmembrane Protein Responsible for Hedgehog Release.
Front Mol Biosci, 8, 2021
|
|
7EKQ
 
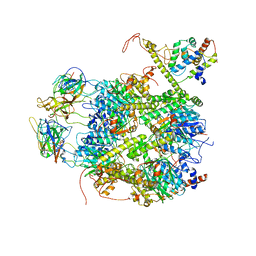 | | CrClpP-S2c | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7EKO
 
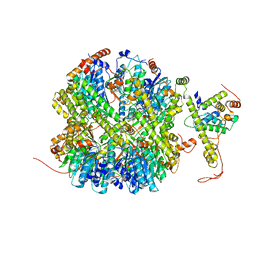 | | CrClpP-S1 | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7DR7
 
 | | bovine 20S immunoproteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2020-12-26 | | Release date: | 2021-01-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of mammalian PA28 alpha beta-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28 alpha beta.
Nat Commun, 12, 2021
|
|
7DMP
 
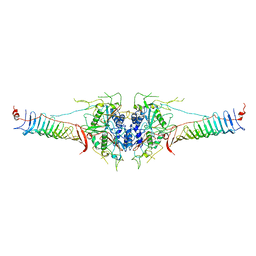 | | Mouse radial spoke complex | | Descriptor: | Radial spoke head 1 homolog, Radial spoke head protein 4 homolog A, Radial spoke head protein 9 homolog | | Authors: | Zheng, W, Cong, Y. | | Deposit date: | 2020-12-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct architecture and composition of mouse axonemal radial spoke head revealed by cryo-EM
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ECY
 
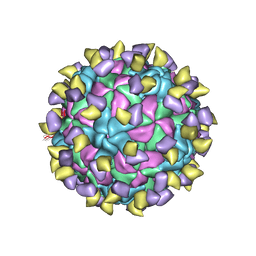 | | EV-D68 in complex with 2H12 Fab (State 3) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7EBR
 
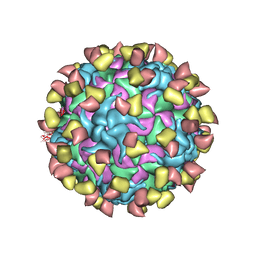 | | EV-D68 in complex with 2H12 Fab (state S2) | | Descriptor: | 2H12 Fab heavy chain, 2H12 Fab light chain, Capsid protein VP1, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-31 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7EBZ
 
 | | EV-D68 in complex with 2H12 Fab (state S1) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-03-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7EC5
 
 | | EV-D68 in complex with 8F12 Fab | | Descriptor: | 8F12 Fab heavy chain, 8F12 Fab light chain, Capsid protein VP1, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-03-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7WD8
 
 | | SARS-CoV-2 Beta spike SD1 in complex with S3H3 Fab | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD0
 
 | | SARS-CoV-2 Beta spike in complex with two S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WDF
 
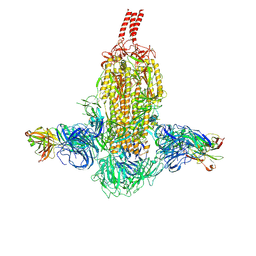 | | SARS-CoV-2 Beta spike in complex with two S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD9
 
 | | SARS-CoV-2 Beta spike in complex with three S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD7
 
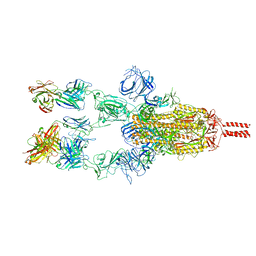 | | SARS-CoV-2 Beta spike in complex with three S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCZ
 
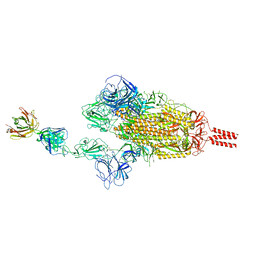 | | SARS-CoV-2 Beta spike in complex with one S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCR
 
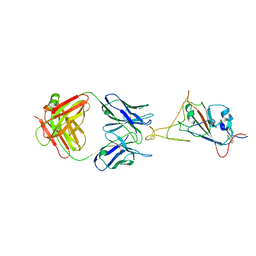 | | RBD-1 of SARS-CoV-2 Beta spike in complex with S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7VX1
 
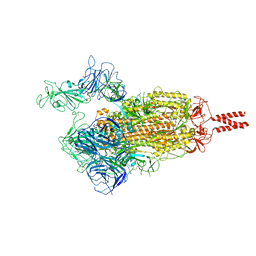 | | SARS-CoV-2 Beta variant spike protein in open state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VX9
 
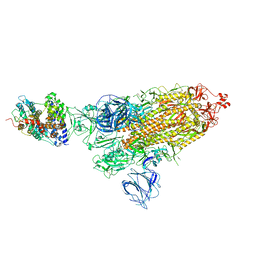 | | SARS-CoV-2 Kappa variant spike protein in complex wth ACE2, state C1 | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXA
 
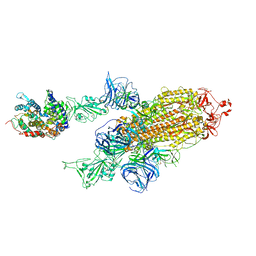 | | SARS-CoV-2 Kappa variant spike protein in complex with ACE2, state C2a | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
