1VR0
 
 | |
1VJL
 
 | |
1VQR
 
 | |
1VR8
 
 | |
1VK3
 
 | |
1VPZ
 
 | |
1VL4
 
 | |
2JZF
 
 | | NMR Conformer closest to the mean coordinates of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
1UWD
 
 | | NMR STRUCTURE OF A PROTEIN WITH UNKNOWN FUNCTION FROM THERMOTOGA MARITIMA (TM0487), WHICH BELONGS TO THE DUF59 FAMILY. | | Descriptor: | HYPOTHETICAL PROTEIN TM0487 | | Authors: | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-02-03 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
2KAF
 
 | | Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus | | Descriptor: | Non-structural protein 3 | | Authors: | Johnson, M.A, Mohanty, B, Pedrini, B, Serrano, P, Chatterjee, A, Herrmann, T, Joseph, J, Saikatendu, K, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding.
J.Mol.Biol., 400, 2010
|
|
2KYS
 
 | | NMR Structure of the SARS Coronavirus Nonstructural Protein Nsp7 in Solution at pH 6.5 | | Descriptor: | Non-structural protein 7 | | Authors: | Johnson, M.A, Jaudzems, K, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the SARS-CoV Nonstructural Protein 7 in Solution at pH 6.5.
J.Mol.Biol., 402, 2010
|
|
3SXK
 
 | |
3SXM
 
 | |
3SXY
 
 | |
3SXZ
 
 | |
9C3G
 
 | |
2ETS
 
 | |
2FNA
 
 | |
2GAZ
 
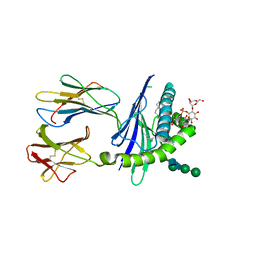 | | Mycobacterial lipoglycan presentation by CD1d | | Descriptor: | (2R)-3-[(HYDROXY{[(2R,3R,5S,6R)-3,4,5-TRIHYDROXY-2,6-BIS(ALPHA-D-MANNOPYRANOSYLOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]PROPAN E-1,2-DIYL DIHEXADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, T-cell surface glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2006-03-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d.
J.Immunol., 177, 2006
|
|
2FIK
 
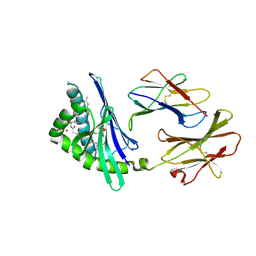 | | Structure of a microbial glycosphingolipid bound to mouse CD1d | | Descriptor: | (2S,3R)-3-HYDROXY-2-(TETRADECANOYLAMINO)OCTADECYL ALPHA-D-GALACTOPYRANOSIDURONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, D, Zajonc, D.M. | | Deposit date: | 2005-12-29 | | Release date: | 2006-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2G36
 
 | |
2FG0
 
 | |
2FEA
 
 | |
2F46
 
 | |
2EVR
 
 | |
