8ER6
 
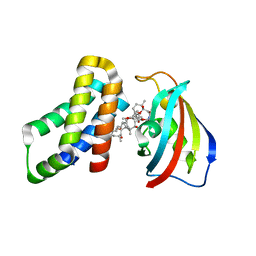 | | FKBP12-FRB in Complex with Compound 11 | | Descriptor: | (3S,5R,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-5,9,27-trihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, 1,2-ETHANEDIOL, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Tomlinson, A.C.A, Yano, J.K. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1-Activated Tumors.
J.Med.Chem., 66, 2023
|
|
8Z02
 
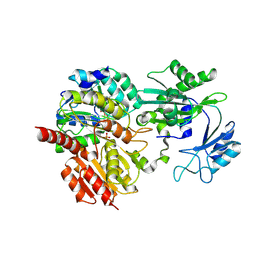 | | CoA bound to human GTP-specific succinyl-CoA synthetase | | Descriptor: | COENZYME A, PHOSPHATE ION, Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, ... | | Authors: | Liu, R.L, Ren, X.L, Li, L.T, Zhang, Y, Huang, H, Zhao, Y.M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis.
Cell Metab., 37, 2025
|
|
8Z03
 
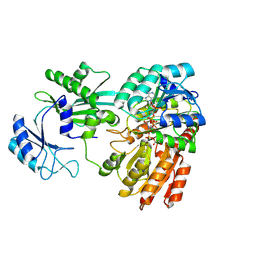 | | Lactate bound to human GTP-specific succinyl-CoA synthetase | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, COENZYME A, MAGNESIUM ION, ... | | Authors: | Liu, R.L, Ren, X.L, Li, L.T, Zhang, Y, Huang, H, Zhao, Y.M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis.
Cell Metab., 37, 2025
|
|
8FB1
 
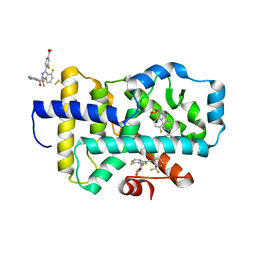 | |
8FB2
 
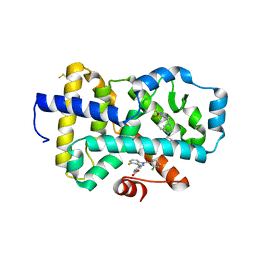 | | HUMAN RETENOID-RELATED ORPHAN RECEPTOR-GAMMA (RORC2) LIGAND-BINDING DOMAIN IN COMPLEX WITH COMPOUND 8 ANDINDAZOLE ACID BOUND IN H12-POCKET | | Descriptor: | (1R,15S)-16-(cyclopropylacetyl)-5-fluoro-20-methyl-9lambda~6~-thia-1,8,16-triazatricyclo[13.3.1.1~3,7~]icosa-3(20),4,6-triene-9,9-dione, 4-[1-(2,6-dichlorobenzoyl)-4-fluoro-1H-indazol-3-yl]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Vajdos, F.F. | | Deposit date: | 2022-11-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Macrocyclic Retinoic Acid Receptor-Related Orphan Receptor C2 Inverse Agonists.
Acs Med.Chem.Lett., 14, 2023
|
|
8FAV
 
 | |
4I6L
 
 | |
8F7C
 
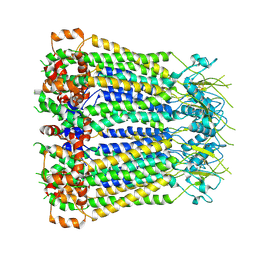 | | Cryo-EM structure of human pannexin 2 | | Descriptor: | Pannexin-2, Soluble cytochrome b562 fusion | | Authors: | He, Z, Yuan, P. | | Deposit date: | 2022-11-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural and functional analysis of human pannexin 2 channel.
Nat Commun, 14, 2023
|
|
6M7L
 
 | | Complex of OxyA with the X-domain from GPA biosynthesis | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase, Putative non-ribosomal peptide synthetase | | Authors: | Greule, A, Izore, T, Tailhades, J, Peschke, M, Schoppet, M, Ahmed, I, Kulik, A, Adamek, M, Ziemert, N, De Voss, J, Stegmann, E, Cryle, M.J. | | Deposit date: | 2018-08-20 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.648297 Å) | | Cite: | Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics.
Nat Commun, 10, 2019
|
|
6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
6AKK
 
 | | Crystal structure of the second Coiled-coil domain of SIKE1 | | Descriptor: | GLYCEROL, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
6AKL
 
 | | Crystal structure of Striatin3 in complex with SIKE1 Coiled-coil domain | | Descriptor: | Striatin-3, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
8XOA
 
 | | VMAT2 complex with MPP+ | | Descriptor: | 1-methyl-4-phenylpyridin-1-ium, Synaptic vesicular amine transporter,Synaptic vesicular amine transporter,transporterA | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-12-31 | | Release date: | 2024-05-08 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
8XO9
 
 | | VMAT2 complex with noradrenaline in cytosol-facing state | | Descriptor: | Noradrenaline, Synaptic vesicular amine transporter,Synaptic vesicular amine transporter,transporter A | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-12-31 | | Release date: | 2024-05-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
8XOB
 
 | | VMAT2 protonated state | | Descriptor: | Synaptic vesicular amine transporter,transporter B | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-12-31 | | Release date: | 2024-05-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
8WQV
 
 | | Fe-O nanocluster of form-VIII in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WPT
 
 | | Truncated mutant (1-171) of ferritin from Ureaplasma diversum | | Descriptor: | CHLORIDE ION, FE (III) ION, Truncated ferritin | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-10-10 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WR0
 
 | | Fe-O nanocluster of form-XII in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WQY
 
 | | Fe-O nanocluster of form-XI in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WPV
 
 | | Truncated mutant (1-171) of ferritin from Ureaplasma diversum soaked in Fe2+ solution for 30min | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-10-10 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WQU
 
 | | Fe-O nanocluster of form-IX in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WQX
 
 | | Fe-O nanocluster of form-X in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
6AKM
 
 | | Crystal structure of SLMAP-SIKE1 complex | | Descriptor: | GLYCEROL, Sarcolemmal membrane-associated protein, Suppressor of IKBKE 1 | | Authors: | Ma, J, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
5YYL
 
 | | Structure of Major Royal Jelly Protein 1 Oligomer | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tian, W, Chen, Z. | | Deposit date: | 2017-12-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architecture of the native major royal jelly protein 1 oligomer.
Nat Commun, 9, 2018
|
|
