7EG7
 
 | | TFIID-based core PIC on SCP promoter | | Descriptor: | DNA (79-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Qi, Y, Hou, H, Wang, X, Wu, Z, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EG8
 
 | | TFIID-based core PIC on PUMA promoter | | Descriptor: | DNA (85-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Qi, Y, Hou, H, Wang, X, Wu, Z, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGA
 
 | | TFIID-based intermediate PIC on PUMA promoter | | Descriptor: | DNA (85-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Wu, Z, Hou, H, Qi, Y, Wang, X, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGC
 
 | | p53-bound TFIID-based holo PIC on HDM2 promoter | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wu, Z, Hou, H, Qi, Y, Wang, X, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
5F5C
 
 | | Crystal Structure of human JMJD2D complexed with KDOPP7 | | Descriptor: | 1,2-ETHANEDIOL, 8-[[(phenylmethyl)amino]methyl]-1~{H}-pyrido[3,4-d]pyrimidin-4-one, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
7CY2
 
 | |
7CZ2
 
 | |
5F5A
 
 | | Crystal Structure of human JMJD2D complexed with KDOAM16 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(furan-2-ylmethylamino)methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F5I
 
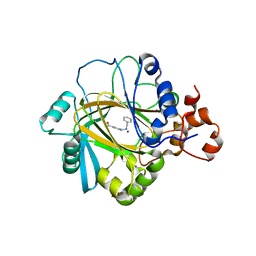 | | Crystal Structure of human JMJD2A complexed with KDOOA011340 | | Descriptor: | 2-[[(phenylmethyl)amino]methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Szykowska, A, Gileadi, C, Johansson, C, England, K, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
7EGG
 
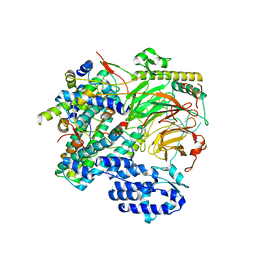 | | TFIID lobe B subcomplex | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 4, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EG9
 
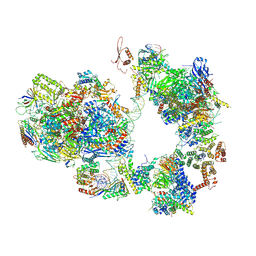 | | TFIID-based intermediate PIC on SCP promoter | | Descriptor: | DNA (79-MER), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGF
 
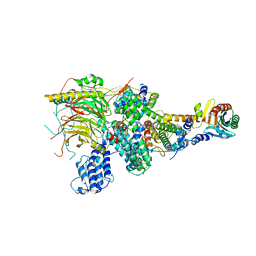 | | TFIID lobe A subcomplex | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 11, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 2021
|
|
7EGH
 
 | | TFIID lobe C subcomplex | | Descriptor: | DNA (45-MER), Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 2, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGI
 
 | | TFIID in rearranged conformation | | Descriptor: | TATA-box-binding protein, Transcription initiation factor IIA subunit 1, Transcription initiation factor IIA subunit 2, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (9.82 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGJ
 
 | | SCP promoter-bound TFIID-TFIIA in post TBP-loading state | | Descriptor: | DNA (74-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (8.64 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGD
 
 | | SCP promoter-bound TFIID-TFIIA in initial TBP-loading state | | Descriptor: | DNA (72-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGE
 
 | | TFIID in canonical conformation | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7CYR
 
 | |
7WQV
 
 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | Authors: | Zha, J, Meng, L, Zhang, X, Li, D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
5UJ2
 
 | | Crystal structure of HCV NS5B genotype 2A JFH-1 isolate with S15G E86Q E87Q C223H V321I mutations and Delta8 neta hairpoin loop deletion in complex with GS-639476 (diphsohate version of GS-9813), Mn2+ and symmetrical primer template 5'-AUAAAUUU | | Descriptor: | (1S)-1-(4-aminoimidazo[2,1-f][1,2,4]triazin-7-yl)-1,4-anhydro-2-deoxy-2-fluoro-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-2-methyl-D-ribitol, CHLORIDE ION, Genome polyprotein, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C, Murakami, E, Rey, A, McGrath, M.E. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a 2'-fluoro-2'-C-methyl C-nucleotide HCV polymerase inhibitor and a phosphoramidate prodrug with favorable properties.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5W45
 
 | | Crystal structure of APOBEC3H | | Descriptor: | APOBEC3H, ZINC ION | | Authors: | Ito, F, Yang, H.J, Xiao, X, Li, S.X, Wolfe, A, Zirkle, B, Arutiunian, V, Chen, X.S. | | Deposit date: | 2017-06-09 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Understanding the Structure, Multimerization, Subcellular Localization and mC Selectivity of a Genomic Mutator and Anti-HIV Factor APOBEC3H.
Sci Rep, 8, 2018
|
|
2GGT
 
 | | Crystal structure of human SCO1 complexed with nickel. | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, SCO1 protein homolog, ... | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Martinelli, M, Palumaa, P, Wang, S. | | Deposit date: | 2006-03-24 | | Release date: | 2006-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5WVR
 
 | | Crystal structure of Osh1 ORD domain in complex with cholesterol | | Descriptor: | CHOLESTEROL, KLLA0C04147p, SULFATE ION | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-12-28 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
2HU7
 
 | | Binding of inhibitors by Acylaminoacyl peptidase | | Descriptor: | ACETYL GROUP, Acylamino-acid-releasing enzyme, GLYCEROL, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
2HU8
 
 | | Binding of inhibitors by Acylaminoacyl peptidase | | Descriptor: | 2-AMINOBENZOIC ACID, Acylamino-acid-releasing enzyme, GLYCINE, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
