1G2U
 
 | | THE STRUCTURE OF THE MUTANT, A172V, OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THERMUS THERMOPHILUS HB8 : ITS THERMOSTABILITY AND STRUCTURE. | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-10-21 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GC8
 
 | | THE CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS 3-ISOPROPYLMALATE DEHYDROGENASE MUTATED AT 172TH FROM ALA TO PHE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-07-27 | | Release date: | 2000-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GC9
 
 | | THE CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS 3-ISOPROPYLMALATE DEHYDROGENASE MUTATED AT 172TH FROM ALA TO GLY | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-07-28 | | Release date: | 2000-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6JTB
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with citrate from Porphyromonas gingivalis (Space) | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
6JTC
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with SH-5 from Porphyromonas gingivalis (Space) | | Descriptor: | 2-(2-azanylethylamino)-5-nitro-benzoic acid, Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
7CAT
 
 | | The NADPH binding site on beef liver catalase | | Descriptor: | CATALASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murthy, M.R.N, Reid III, T.J, Sicignano, A, Tanaka, N, Fita, I, Rossmann, M.G. | | Deposit date: | 1984-11-15 | | Release date: | 1985-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The NADPH binding site on beef liver catalase.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
1IPD
 
 | | THREE-DIMENSIONAL STRUCTURE OF A HIGHLY THERMOSTABLE ENZYME, 3-ISOPROPYLMALATE DEHYDROGENASE OF THERMUS THERMOPHILUS AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | Authors: | Imada, K, Sato, M, Tanaka, N, Katsube, Y, Matsuura, Y, Oshima, T. | | Deposit date: | 1992-01-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
J.Mol.Biol., 222, 1991
|
|
1IOP
 
 | | INCORPORATION OF A HEMIN WITH THE SHORTEST ACID SIDE-CHAINS INTO MYOGLOBIN | | Descriptor: | 6,7-DICARBOXYL-1,2,3,4,5,8-HEXAMETHYLHEMIN, CYANIDE ION, MYOGLOBIN, ... | | Authors: | Igarashi, N, Neya, S, Funasaki, N, Tanaka, N. | | Deposit date: | 1997-12-12 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of 6,7-dicarboxyheme-substituted myoglobin
Biochemistry, 37, 1998
|
|
1WTN
 
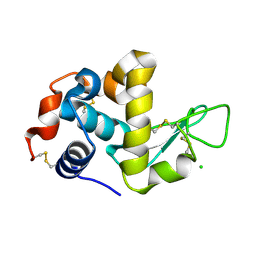 | | The structure of HEW Lysozyme Orthorhombic Crystal Growth under a High Magnetic Field | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Saijo, S, Yamada, Y, Sato, T, Tanaka, N, Matsui, T, Sazaki, G, Nakajima, K, Matsuura, Y. | | Deposit date: | 2004-11-25 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural consequences of hen egg-white lysozyme orthorhombic crystal growth in a high magnetic field: validation of X-ray diffraction intensity, conformational energy searching and quantitative analysis of B factors and mosaicity.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ITK
 
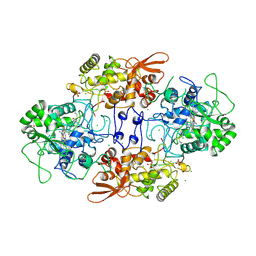 | | Crystal structure of catalase-peroxidase from Haloarcula marismortui | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Yamada, Y, Fujiwara, T, Sato, T, Igarashi, N, Tanaka, N. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of catalase-peroxidase from Haloarcula marismortui.
Nat.Struct.Biol., 9, 2002
|
|
1J2T
 
 | | Creatininase Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, ZINC ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1J2U
 
 | | Creatininase Zn | | Descriptor: | SULFATE ION, ZINC ION, creatinine amidohydrolase | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1IDM
 
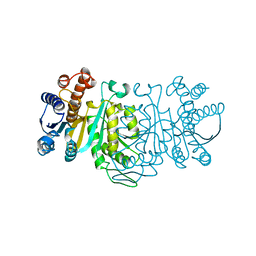 | | 3-ISOPROPYLMALATE DEHYDROGENASE, LOOP-DELETED CHIMERA | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Sakurai, M, Ohzeki, M, Moriyama, H, Sato, M, Tanaka, N. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a loop-deleted variant of 3-isopropylmalate dehydrogenase from Thermus thermophilus: an internal reprieve tolerance mechanism.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1WNI
 
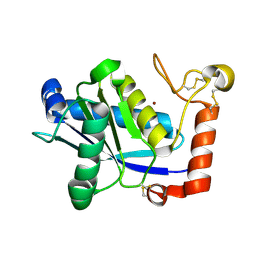 | | Crystal Structure of H2-Proteinase | | Descriptor: | Trimerelysin II, ZINC ION | | Authors: | Kumasaka, T, Yamamoto, M, Moriyama, H, Tanaka, N, Sato, M, Katsube, Y, Yamakawa, Y, Omori-Satoh, T, Iwanaga, S, Ueki, T. | | Deposit date: | 2004-08-04 | | Release date: | 2004-08-17 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of H2-proteinase from the venom of Trimeresurus flavoviridis.
J.Biochem., 119, 1996
|
|
1XER
 
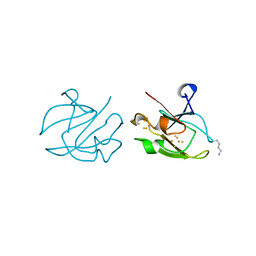 | | STRUCTURE OF FERREDOXIN | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, ZINC ION | | Authors: | Fujii, T, Hata, Y, Moriyama, H, Wakagi, T, Tanaka, N, Oshima, T. | | Deposit date: | 1996-08-28 | | Release date: | 1997-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding centre in thermoacidophilic archaeal ferredoxins.
Nat.Struct.Biol., 3, 1996
|
|
1XAC
 
 | |
1XAD
 
 | |
1VEC
 
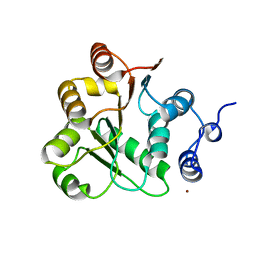 | | Crystal structure of the N-terminal domain of rck/p54, a human DEAD-box protein | | Descriptor: | ATP-dependent RNA helicase p54, L(+)-TARTARIC ACID, ZINC ION | | Authors: | Hogetsu, K, Matsui, T, Yukihiro, Y, Tanaka, M, Sato, T, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-03-29 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insight of human DEAD-box protein rck/p54 into its substrate recognition with conformational changes
Genes Cells, 11, 2006
|
|
7DKD
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Asn-Tyr | | Descriptor: | ASPARAGINE, Dipeptidyl-peptidase, GLYCEROL, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKC
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Tyr-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, TYROSINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKE
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Phe-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, PHENYLALANINE, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKB
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Val-Tyr | | Descriptor: | Dipeptidyl-peptidase, TYROSINE, VALINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
1VBU
 
 | | Crystal structure of native xylanase 10B from Thermotoga maritima | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Ihsanawati, Kumasaka, T, Kaneko, T, Nakamura, S, Tanaka, N. | | Deposit date: | 2004-03-02 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8
Proteins, 61, 2005
|
|
1VBR
 
 | | Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose | | Descriptor: | ACETIC ACID, alpha-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase B | | Authors: | Ihsanawati, Kumasaka, T, Kaneko, T, Nakamura, S, Tanaka, N. | | Deposit date: | 2004-03-02 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8
Proteins, 61, 2005
|
|
1WQS
 
 | | Crystal structure of Norovirus 3C-like protease | | Descriptor: | 3C-like protease, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Nakamura, K, Someya, Y, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-10-01 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A norovirus protease structure provides insights into active and substrate binding site integrity
J.Virol., 79, 2005
|
|
