9HAL
 
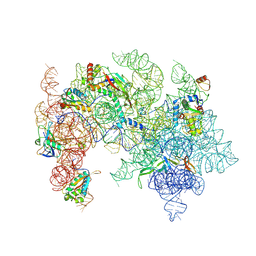 | | Pooled 50S subunit d126_(L29)-/(L22)- precursor states supplemented with Api137 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Large ribosomal subunit protein bL17, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-04 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.49 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9HA6
 
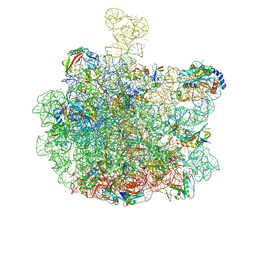 | | mature 50S subunit supplemented with Api137 | | Descriptor: | 23S ribosomal rRNA, 50S ribosomal protein L16, 50S ribosomal protein L3, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-01 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3L
 
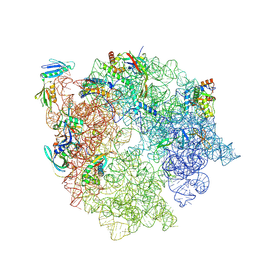 | | 50S subunit precursor C_(L29)-/(L22)- | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Large ribosomal subunit protein bL17, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (5.84 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3Z
 
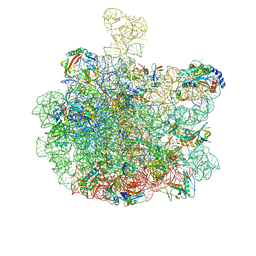 | | mature 50S subunit | | Descriptor: | 23S ribosomal rRNA, 50S ribosomal protein L16, 50S ribosomal protein L3, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3W
 
 | | 50S subunit precursor C-CP_H68 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, 5S ribosomal RNA, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (5.38 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9H3M
 
 | | 50S subunit precursor C_(L22)- | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Large ribosomal subunit protein bL17, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.41 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9HA5
 
 | | Pooled 50S subunit C_L2 precursor states supplemented with Api137 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Apidaecins type 22, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-01 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9HAM
 
 | | C_(L29)-/(L22)- precursor supplemented with Api137 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Large ribosomal subunit protein bL17, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-04 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (5.06 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9HAI
 
 | | Pooled 50S subunit C-CP_L2-L28 precursor states supplemented with Api137 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Apidaecins type 22, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-04 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
9HA1
 
 | | Pooled 50S subunit C_(L22)- precursor states supplemented with Api137 - Canonical PET exit Api137 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Apidaecins type 137, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2024-11-01 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | The proline-rich antimicrobial peptide Api137 disrupts large ribosomal subunit assembly and induces misfolding.
Nat Commun, 16, 2025
|
|
2IWH
 
 | | Structure of yeast Elongation Factor 3 in complex with ADPNP | | Descriptor: | ELONGATION FACTOR 3A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
9FL7
 
 | |
2IW3
 
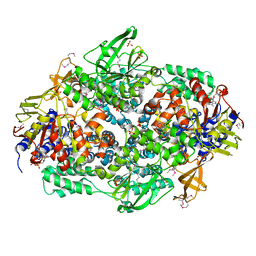 | | Elongation Factor 3 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELONGATION FACTOR 3A, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
2IX3
 
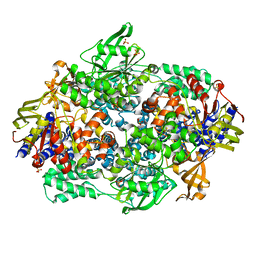 | | Structure of yeast Elongation Factor 3 | | Descriptor: | ELONGATION FACTOR 3, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
7N30
 
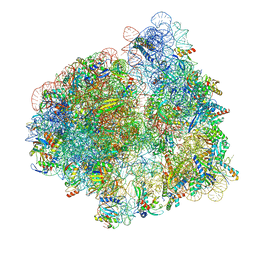 | | Elongating 70S ribosome complex in a hybrid-H2* pre-translocation (PRE-H2*) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-30 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N2C
 
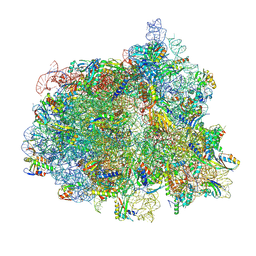 | | Elongating 70S ribosome complex in a fusidic acid-stalled intermediate state of translocation bound to EF-G(GDP) (INT2) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N31
 
 | | Elongating 70S ribosome complex in a post-translocation (POST) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N1P
 
 | | Elongating 70S ribosome complex in a classical pre-translocation (PRE-C) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, S.K, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N2U
 
 | | Elongating 70S ribosome complex in a hybrid-H1 pre-translocation (PRE-H1) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N2V
 
 | | Elongating 70S ribosome complex in a spectinomycin-stalled intermediate state of translocation bound to EF-G in an active, GTP conformation (INT1) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
4D67
 
 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 28S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em Structures of Ribosomal 80S Complexes with Termination Factors and Cricket Paralysis Virus Ires Reveal the Ires in the Translocated State
Mol.Cell, 57, 2015
|
|
4D61
 
 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4D5N
 
 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | CRICKET PARALYSIS VIRUS IRES RNA, EUKARYOTIC PEPTIDE CHAIN RELEASE FACTOR SUBUNIT 1 | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-06 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4D5L
 
 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 18S RRNA 2, 40S RIBOSOMAL PROTEIN ES1, 40S RIBOSOMAL PROTEIN ES10, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
6GBZ
 
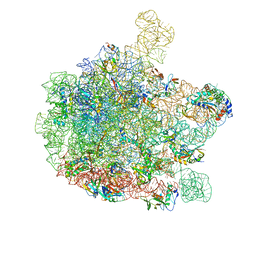 | | 50S ribosomal subunit assembly intermediate state 5 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
