7CJU
 
 | |
3UNC
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase to 1.65A Resolution | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
3UNI
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with NADH Bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-HYDROXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
3UNA
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with NAD Bound | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
5ZXW
 
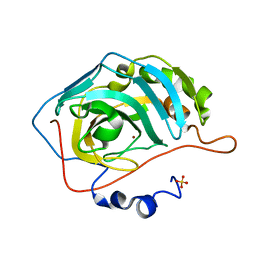 | | Crystal structure of human carbonic anhydrase II crystallized by ammonium sulfate | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION | | Authors: | Kitahara, M, Fudo, S, Yoneda, T, Nukaga, M, Hoshino, T. | | Deposit date: | 2018-05-22 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.316 Å) | | Cite: | Anisotropic Distribution of Ammonium Sulfate Ions in Protein Crystallization
Cryst.Growth Des., 19, 2019
|
|
5ZZE
 
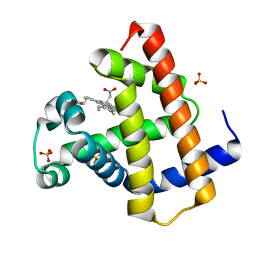 | | Crystal structure of horse myoglobin crystallized by ammonium sulfate | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kitahara, M, Fudo, S, Yoneda, T, Nukaga, M, Hoshino, T. | | Deposit date: | 2018-06-01 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Anisotropic Distribution of Ammonium Sulfate Ions in Protein Crystallization
Cryst.Growth Des., 19, 2019
|
|
6A10
 
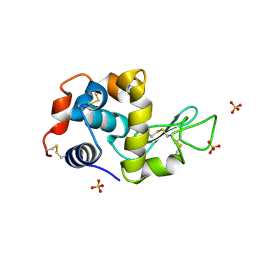 | | Crystal structure of hen egg white lysozyme crystallized by ammonium sulfate | | Descriptor: | Lysozyme C, SULFATE ION | | Authors: | Kitahara, M, Fudo, S, Yoneda, T, Nukaga, M, Hoshino, T. | | Deposit date: | 2018-06-06 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Anisotropic Distribution of Ammonium Sulfate Ions in Protein Crystallization
Cryst.Growth Des., 19, 2019
|
|
4KT8
 
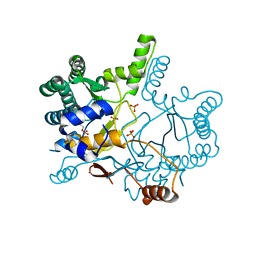 | | The complex structure of Rv3378c-Y51FY90F with substrate, TPP | | Descriptor: | (2E)-3-methyl-5-[(1R,2S,8aS)-1,2,5,5-tetramethyl-1,2,3,5,6,7,8,8a-octahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
7XJ5
 
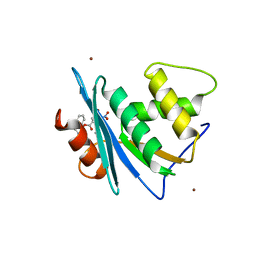 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XIS
 
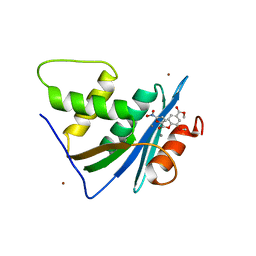 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | (2-methoxy-4-methoxycarbonyl-phenyl) 5-nitrofuran-2-carboxylate, MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XIU
 
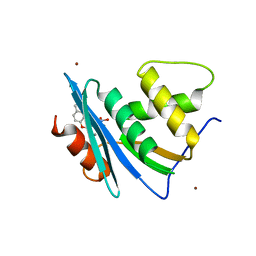 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XIT
 
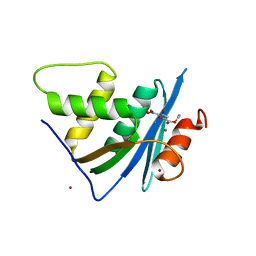 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XJ7
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XJ4
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, S-[5-[(E)-2-phenylethenyl]-1,3,4-oxadiazol-2-yl] 5-nitrothiophene-2-carbothioate, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XGQ
 
 | |
7XGC
 
 | |
2ZRU
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Isopentenyl-diphosphate delta-isomerase | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
2ZRX
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with FMN and DMAPP. | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Isopentenyl-diphosphate delta-isomerase, ... | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
2ZRZ
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with reduced FMN and DMAPP | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, DIMETHYLALLYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, ... | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
2ZRV
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with reduced FMN. | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Isopentenyl-diphosphate delta-isomerase | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
2ZRY
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with reduced FMN and IPP. | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, ... | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
1V97
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase FYX-051 bound form | | Descriptor: | 4-(5-PYRIDIN-4-YL-1H-1,2,4-TRIAZOL-3-YL)PYRIDINE-2-CARBONITRILE, ACETIC ACID, CALCIUM ION, ... | | Authors: | Okamoto, K, Matsumoto, K, Hille, R, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2004-01-21 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The crystal structure of xanthine oxidoreductase during catalysis: Implications for reaction mechanism and enzyme inhibition.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2ZRW
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with FMN and IPP. | | Descriptor: | FLAVIN MONONUCLEOTIDE, ISOPENTYL PYROPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, ... | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
2E1Q
 
 | | Crystal Structure of Human Xanthine Oxidoreductase mutant, Glu803Val | | Descriptor: | 2-HYDROXYBENZOIC ACID, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Yamaguchi, Y, Matsumura, T, Ichida, K, Okamoto, K, Nishino, T. | | Deposit date: | 2006-10-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate
J.Biochem.(Tokyo), 141, 2007
|
|
1VDV
 
 | | Bovine Milk Xanthine Dehydrogenase Y-700 Bound Form | | Descriptor: | 1-[3-CYANO-4-(NEOPENTYLOXY)PHENYL]-1H-PYRAZOLE-4-CARBOXYLIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Fukunari, A, Okamoto, K, Nishino, T, Eger, B.T, Pai, E.F, Kamezawa, M, Yamada, I, Kato, N. | | Deposit date: | 2004-03-25 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Y-700 [1-[3-Cyano-4-(2,2-dimethylpropoxy)phenyl]-1H-pyrazole-4-carboxylic acid]: a potent xanthine oxidoreductase inhibitor with hepatic excretion
J.Pharmacol.Exp.Ther., 311, 2004
|
|
