7MIO
 
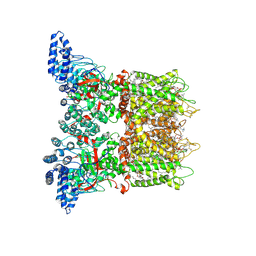 | | Mouse TRPV3 in cNW11 nanodiscs, open state at 42 degrees Celsius | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MIK
 
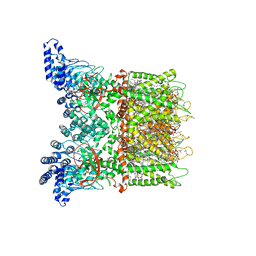 | | Mouse TRPV3 in MSP2N2 nanodiscs, closed state at 42 degrees Celsius | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MIN
 
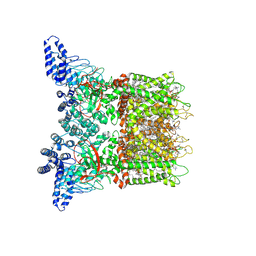 | | Mouse TRPV3 in cNW11 nanodiscs, closed state at 42 degrees Celsius | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6RTZ
 
 | |
6RU0
 
 | |
2K9P
 
 | |
4BXU
 
 | | Structure of Pex14 in complex with Pex5 LVxEF motif | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Kooshapur, H, Meyer, H.N, Madl, T, Sattler, M. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Novel Pex14 Interacting Site of Human Pex5 is Critical for Matrix Protein Import Into Peroxisomes.
J.Biol.Chem., 289, 2014
|
|
4IUK
 
 | |
4IUH
 
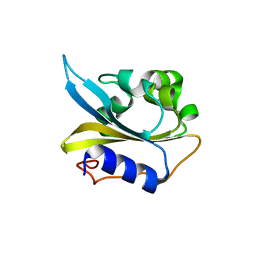 | |
9EOK
 
 | |
9FKD
 
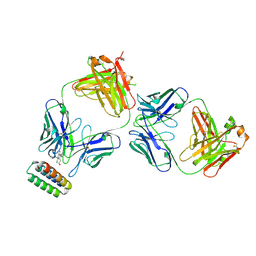 | |
9EOJ
 
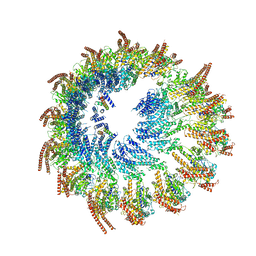 | | Vertebrate microtubule-capping gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component, Gamma-tubulin complex component 3 homolog, Gamma-tubulin complex component 6, ... | | Authors: | Vermeulen, B.J.A, Pfeffer, S. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | gamma-TuRC asymmetry induces local protofilament mismatch at the RanGTP-stimulated microtubule minus end.
Embo J., 43, 2024
|
|
8S1X
 
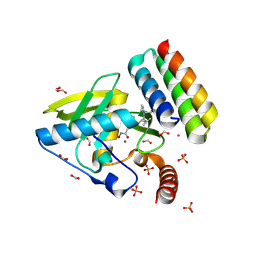 | |
7ANZ
 
 | | Structure of the Candida albicans gamma-Tubulin Small Complex | | Descriptor: | Spindle pole body component, Tubulin gamma chain | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2020-10-13 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The cryo-EM structure of a gamma-TuSC elucidates architecture and regulation of minimal microtubule nucleation systems.
Nat Commun, 11, 2020
|
|
8AT2
 
 | | Structure of the augmin TIII subcomplex | | Descriptor: | HAUS augmin like complex subunit 4 L homeolog, HAUS augmin-like complex subunit 1, HAUS augmin-like complex subunit 3, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The augmin complex architecture reveals structural insights into microtubule branching.
Nat Commun, 13, 2022
|
|
8AT4
 
 | | Structure of the augmin holocomplex in closed conformation | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, HAUS augmin like complex subunit 4 L homeolog, HAUS augmin like complex subunit 6 L homeolog, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (33 Å) | | Cite: | The augmin complex architecture reveals structural insights into microtubule branching.
Nat Commun, 13, 2022
|
|
8AT3
 
 | | Structure of the augmin holocomplex in open conformation | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, HAUS augmin like complex subunit 4 L homeolog, HAUS augmin like complex subunit 6 L homeolog, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (33 Å) | | Cite: | The augmin complex architecture reveals structural insights into microtubule branching.
Nat Commun, 13, 2022
|
|
6G70
 
 | | Structure of murine Prpf39 | | Descriptor: | Pre-mRNA-processing factor 39 | | Authors: | De Bortoli, F.D, Loll, B, Wahl, M, Heyd, F. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Increased versatility despite reduced molecular complexity: evolution, structure and function of metazoan splicing factor PRPF39.
Nucleic Acids Res., 47, 2019
|
|
5MM3
 
 | | Unstructured MamC magnetite-binding protein located between two helices. | | Descriptor: | Sugar ABC transporter substrate-binding protein,Magnetosome protein MamC,Sugar ABC transporter substrate-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nudelman, H, Zarivach, R. | | Deposit date: | 2016-12-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The importance of the helical structure of a MamC-derived magnetite-interacting peptide for its function in magnetite formation.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7PIE
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN068 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-5-[3-[(2R)-morpholin-2-yl]phenyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.427 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PIG
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN088 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-azanyl-2-chloranyl-3-[(Z)-piperidin-3-ylidenemethyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PID
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN060 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-5-[3-[(3R)-morpholin-3-yl]phenyl]benzenecarbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PIF
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN086 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[3-[4-(aminomethyl)oxan-4-yl]phenyl]-2-azanyl-benzenecarbonitrile, CHLORIDE ION, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PIH
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN093 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[3-[[(3~{R})-3-azanylpyrrolidin-1-yl]methyl]phenyl]-4~{H}-isoquinolin-1-one, CHLORIDE ION, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
7PNS
 
 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN081 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-6-[5-[(dimethylamino)methyl]-2-fluoranyl-phenyl]-1H-indole-3-carbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
