7ZFC
 
 | | SARS-CoV-2 Beta RBD in complex with nanobody C1, Omi-18 and Omi-31 Fabs | | Descriptor: | Nanobody C1, Omi-18 heavy chain, Omi-18 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF9
 
 | | SARS-CoV-2 Omicron BA.2 RBD in complex with COVOX-150 Fab (P21) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-150 heavy chain, COVOX-150 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZRC
 
 | | OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-38 Fab Heavy Chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-04 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF5
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-12 and Beta-54 Fabs | | Descriptor: | Beta-54 heavy chain, Beta-54 light chain, Omi-12 heavy chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5.32 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF3
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-3 and EY6A Fabs | | Descriptor: | EY6A heavy chain, EY6A light chain, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZR7
 
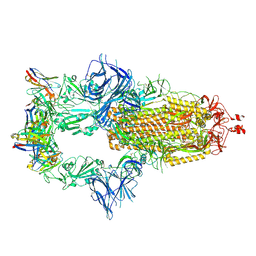 | | OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-42 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFB
 
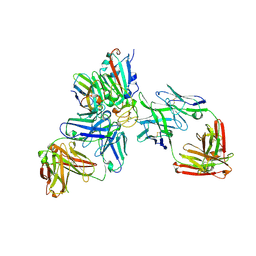 | | SARS-CoV-2 Omicron RBD in complex with nanobody C1, Omi-18 and Omi-31 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody C1, Omi-18 heavy chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF4
 
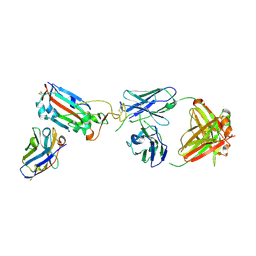 | | SARS-CoV-2 Omicron RBD in complex with Omi-9 Fab and nanobody F2 | | Descriptor: | Nanobody F2, Omi-9 heavy chain, Omi-9 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF7
 
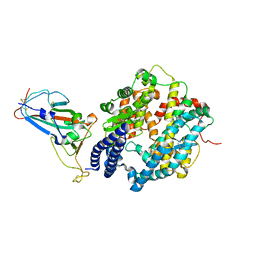 | | SARS-CoV-2 Omicron BA.2 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZR8
 
 | | OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-38 Fab light chain, Omi-38 fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZXU
 
 | | SARS-CoV-2 Omicron BA.4/5 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-27 heavy chain, Beta-27 light chain, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-23 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum.
Cell, 185, 2022
|
|
7ZFA
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-6 and COVOX-150 Fabs | | Descriptor: | COVOX-150 heavy chain, COVOX-150 light chain, Omi-6 heavy chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.24 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZMY
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN0
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN3
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the L state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN9
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 7.0 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNB
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 5.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNA
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 5.2 in the presence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZND
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 7.6 in the absence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN8
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 7.0 in the presence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNC
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 7.6 in the absence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNE
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 8.2 at room temperature, 7.5-ms-long snapshots | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNG
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 8.2 at room temperature, 500-mks-long snapshots | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNH
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the activated state at pH 8.2 at room temperature, 250-750-mks-snapshot | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNI
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the activated state at pH 8.2 at room temperature, 7.5-15-ms-snapshot | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
