8TCC
 
 | |
4F7E
 
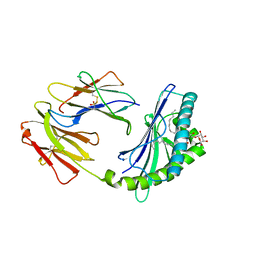 | | Crystal structure of bovine CD1d with bound C16:0-alpha-galactosyl ceramide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Wang, J, Zajonc, D.M. | | Deposit date: | 2012-05-15 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Bovine CD1d Reveal Altered αGalCer Presentation and a Restricted A' Pocket Unable to Bind Long-Chain Glycolipids.
Plos One, 7, 2012
|
|
4I6S
 
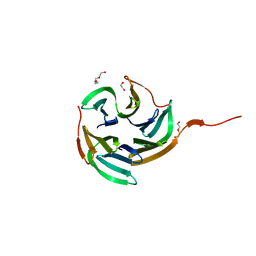 | | Structure of RSL mutant W76A in complex with L-fucose | | Descriptor: | 1,2-ETHANEDIOL, Putative fucose-binding lectin protein, TETRAETHYLENE GLYCOL, ... | | Authors: | Audfray, A, Arnaud, J, Varrot, A, Imberty, A. | | Deposit date: | 2012-11-30 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Reduction of lectin valency drastically changes glycolipid dynamics in membranes but not surface avidity
Acs Chem.Biol., 8, 2013
|
|
4F7C
 
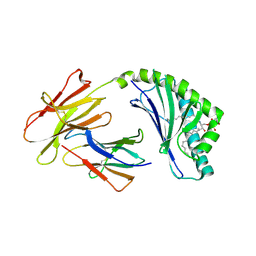 | | Crystal structure of bovine CD1d with bound C12-di-sulfatide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CD1D antigen, ... | | Authors: | Wang, J, Zajonc, D.M. | | Deposit date: | 2012-05-15 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | Crystal Structures of Bovine CD1d Reveal Altered αGalCer Presentation and a Restricted A' Pocket Unable to Bind Long-Chain Glycolipids.
Plos One, 7, 2012
|
|
7NR1
 
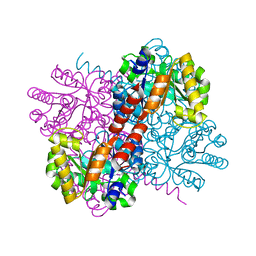 | | Crystal structure of holo-S116A mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 | | Descriptor: | HpcH/HpaI aldolase, MAGNESIUM ION | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-03-02 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7NNK
 
 | | Crystal structure of S116A mutant of hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, HpcH/HpaI aldolase, MAGNESIUM ION | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-02-25 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7SMG
 
 | | Crystal structure of a (p)ppApp hydrolase from Bacteroides caccae | | Descriptor: | (p)ppApp hydrolase, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Ahmad, S, Alexei, A.G, Tsang, K.K, Trilesky, S, Kim, Y, Whitney, J.C. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-09 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a broadly conserved family of enzymes that hydrolyze (p)ppApp.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7JX2
 
 | |
7JVG
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 1-arachidonoylglycerol | | Descriptor: | (2S)-2,3-dihydroxypropyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Golczak, M. | | Deposit date: | 2020-08-21 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
7JVY
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 2-arachidonylglyceryl ether | | Descriptor: | 2-{[(5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraen-1-yl]oxy}propane-1,3-diol, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Banarjee, S, Golczak, M. | | Deposit date: | 2020-08-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
7JWR
 
 | |
7JWD
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 2-linoleoylglycerol | | Descriptor: | 1,3-dihydroxypropan-2-yl (9Z,12Z)-octadeca-9,12-dienoate, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Banarjee, S, Golczak, M. | | Deposit date: | 2020-08-25 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35000193 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
7K3I
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 2-lauroylglycerol | | Descriptor: | 1,3-dihydroxypropan-2-yl dodecanoate, Retinol-binding protein 2 | | Authors: | Adams, C, Silvaroli, J.A, Banarjee, S, Golczak, M. | | Deposit date: | 2020-09-11 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
7JZ5
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 1-arachodonoyl-1-thio-glycerol | | Descriptor: | Retinol-binding protein 2, S-[(2R)-2,3-dihydroxypropyl] (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenethioate | | Authors: | Silvaroli, J.A, Banarjee, S, Golczak, M. | | Deposit date: | 2020-09-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.567 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
8ADQ
 
 | | Crystal structure of holo-SwHPA-Mg (hydroxy ketone aldolase) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate and D-Glyceraldehyde | | Descriptor: | 3-HYDROXYPYRUVIC ACID, BROMIDE ION, D-Glyceraldehyde, ... | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5CE4
 
 | | High Resolution X-Ray and Neutron diffraction structure of H-FABP | | Descriptor: | Fatty acid-binding protein, heart, OLEIC ACID | | Authors: | Podjarny, A.D, Howard, E.I, Blakeley, M.P, Guillot, B. | | Deposit date: | 2015-07-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (0.98 Å), X-RAY DIFFRACTION | | Cite: | High-resolution neutron and X-ray diffraction room-temperature studies of an H-FABP-oleic acid complex: study of the internal water cluster and ligand binding by a transferred multipolar electron-density distribution.
Iucrj, 3, 2016
|
|
5EJS
 
 | | Structure of Dictyostelium Discoideum Myosin VII MyTH4-FERM MF2 domain, mutant 1 | | Descriptor: | Myosin-I heavy chain | | Authors: | Planelles-Herrero, V.J, Sirkia, H, Sourigues, Y, Titus, M.A, Houdusse, A. | | Deposit date: | 2015-11-02 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Myosin MyTH4-FERM structures highlight important principles of convergent evolution.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EJY
 
 | |
5EJQ
 
 | | Structure of Dictyostelium Discoideum Myosin VII MyTH4-FERM MF1 domain, mutant 2 | | Descriptor: | GLUTAMIC ACID, GLYCINE, Myosin-I heavy chain, ... | | Authors: | Planelles-Herrero, V.J, Sirkia, H, Sourigues, Y, Titus, M.A, Houdusse, A. | | Deposit date: | 2015-11-02 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Myosin MyTH4-FERM structures highlight important principles of convergent evolution.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7APS
 
 | | The Fk1 domain of FKBP51 in complex with (2S)-2-((1S,5R,6R)-10-((3,5-dichlorophenyl)sulfonyl)-2-oxo-5-vinyl-3,10-diazabicyclo[4.3.1]decan-3-yl)propanoic acid | | Descriptor: | (2~{S})-2-[(1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethyl-2-oxidanylidene-3,10-diazabicyclo[4.3.1]decan-3-yl]propanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Kolos, M.J, Pomplun, S, Riess, B, Purder, P, Voll, M.A, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
7APT
 
 | | The Fk1 domain of FKBP51 in complex with ((1S,5S,6R)-10-((3,5-dichlorophenyl)sulfonyl)-2-oxo-5-vinyl-3,10-diazabicyclo[4.3.1]decan-3-yl)acetic acid | | Descriptor: | 2-[(1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-2-oxidanylidene-3,10-diazabicyclo[4.3.1]decan-3-yl]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Kolos, M.J, Pomplun, S, Riess, B, Purder, P, Voll, M.A, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
7APQ
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-(1,3-benzothiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Kolos, M.J, Pomplun, S, Riess, B, Purder, P, Voll, M.A, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
7APW
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, A.M, Kolos, J.M, Pomplun, S, Riess, B, Purder, P, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-20 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
2LIF
 
 | | Solution Structure of KKGF | | Descriptor: | Core protein p21 | | Authors: | Montserret, R, Penin, F. | | Deposit date: | 2011-08-29 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of hepatitis C virus core-e1 signal Peptide and requirements for cleavage of the genotype 3a signal sequence by signal Peptide peptidase.
J.Virol., 86, 2012
|
|
