3KTI
 
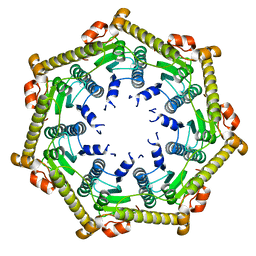
 | | Structure of ClpP in complex with ADEP1 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ATP-dependent Clp protease proteolytic subunit, Acyldepsipeptide 1, ... | | Authors: | Lee, B.-G, Brotz-Oesterhelt, H, Song, H.K. | | Deposit date: | 2009-11-25 | | Release date: | 2010-03-23 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KTK
 
 | |
3KTH
 
 | |
3KTG
 
 | |
3KU7
 
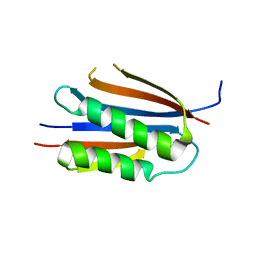 | | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor | | Descriptor: | Cell division topological specificity factor | | Authors: | Kang, G.B, Song, H.E, Kim, M.K, Eom, S.H. | | Deposit date: | 2009-11-26 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor
Mol.Microbiol., 76, 2010
|
|
2DS6
 
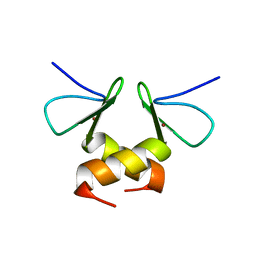 | | Structure of the ZBD in the tetragonal crystal form | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2DS8
 
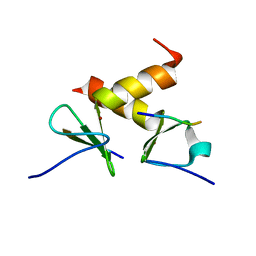 | | Structure of the ZBD-XB complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, SspB-tail peptide, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Kim, H.W, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2DS5
 
 | | Structure of the ZBD in the orthorhomibic crystal from | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Song, H.K, Park, E.Y, Lee, B.G, Hong, S.B. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2DS7
 
 | | Structure of the ZBD in the hexagonal crystal form | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2GX2
 
 | | Crystal structural and functional analysis of GFP-like fluorescent protein Dronpa | | Descriptor: | MAGNESIUM ION, fluorescent protein Dronpa | | Authors: | Hwang, K.Y, Nam, K.-H, Park, S.-Y, Sugiyama, K. | | Deposit date: | 2006-05-08 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the photoswitchable fluorescent protein Dronpa-C62S
Biochem.Biophys.Res.Commun., 354, 2007
|
|
5J45
 
 | |
1L0Q
 
 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
3BWE
 
 | | Crystal structure of aggregated form of DJ1 | | Descriptor: | PHOSPHATE ION, Protein DJ-1 | | Authors: | Cha, S.S. | | Deposit date: | 2008-01-09 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of filamentous aggregates of human DJ-1 formed in an inorganic phosphate-dependent manner.
J.Biol.Chem., 283, 2008
|
|
3RUJ
 
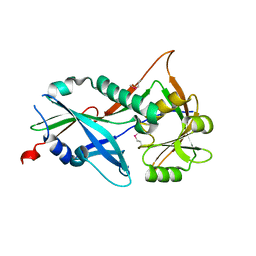 | |
3RUI
 
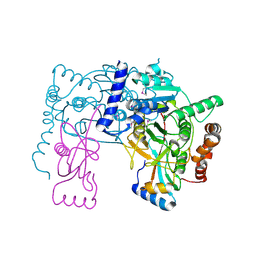 | | Crystal structure of Atg7C-Atg8 complex | | Descriptor: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Hong, S.B, Kim, B.W, Song, H.K. | | Deposit date: | 2011-05-05 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2GX0
 
 | |
4IM2
 
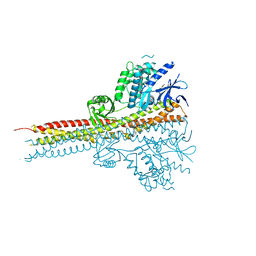 | | Structure of Tank-Binding Kinase 1 | | Descriptor: | CHLORIDE ION, N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Tu, D, Eck, M.J. | | Deposit date: | 2013-01-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | Structure and ubiquitination-dependent activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4IM0
 
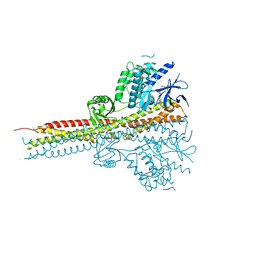 | | Structure of Tank-Binding Kinase 1 | | Descriptor: | N-{3-[(5-cyclopropyl-2-{[3-(morpholin-4-ylmethyl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Tu, D, Eck, M.J. | | Deposit date: | 2013-01-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4001 Å) | | Cite: | Structure and ubiquitination-dependent activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4IM3
 
 | | Structure of Tank-Binding Kinase 1 | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, ... | | Authors: | Tu, D, Eck, M.J. | | Deposit date: | 2013-01-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.342 Å) | | Cite: | Structure and ubiquitination-dependent activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
5VKZ
 
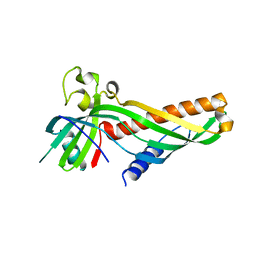 | | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Egea, P.F, AhYoung, A.P, Lu, B, Tan, H.R, Cascio, D. | | Deposit date: | 2017-04-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
6RBN
 
 | |
6RAP
 
 | |
6RAO
 
 | |
6RGL
 
 | |
6RBK
 
 | |
