2A9E
 
 | |
1ZUA
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Tolrestat | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT | | Authors: | Gallego, O, Ruiz, F.X, Ardevol, A, Dominguez, M, Alvarez, R, de Lera, A.R, Rovira, C, Farres, J, Fita, I, Pares, X. | | Deposit date: | 2005-05-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2BIS
 
 | | Structure of glycogen synthase from Pyrococcus abyssi | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLGA GLYCOGEN SYNTHASE, GLYCEROL, ... | | Authors: | Horcajada, C, Guinovart, J.J, Fita, I, Ferrer, J.C. | | Deposit date: | 2005-01-25 | | Release date: | 2005-11-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of an Archaeal Glycogen Synthase: Insights Into Oligomerisation and Substrate Binding of Eukaryotic Glycogen Synthases.
J.Biol.Chem., 281, 2006
|
|
2BFW
 
 | | Structure of the C domain of glycogen synthase from Pyrococcus abyssi | | Descriptor: | ACETATE ION, GLGA GLYCOGEN SYNTHASE, SULFATE ION | | Authors: | Horcajada, C, Guinovart, J.J, Fita, I, Ferrer, J.C. | | Deposit date: | 2004-12-15 | | Release date: | 2005-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Archaeal Glycogen Synthase: Insights Into Oligomerisation and Substrate Binding of Eukaryotic Glycogen Synthases.
J.Biol.Chem., 281, 2006
|
|
2BTY
 
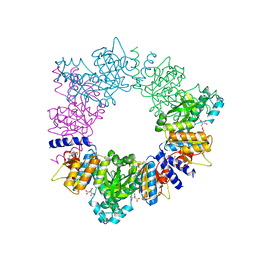 | | Acetylglutamate kinase from Thermotoga maritima complexed with its inhibitor arginine | | Descriptor: | ACETYLGLUTAMATE KINASE, ARGININE, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Gil-Ortiz, F, Fernandez-Murga, M.L, Fita, I, Rubio, V. | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Bases of Feed-Back Control of Arginine Biosynthesis, Revealed by the Structure of Two Hexameric N-Acetylglutamate Kinases, from Thermotoga Maritima and Pseudomonas Aeruginosa
J.Mol.Biol., 356, 2006
|
|
2J8Z
 
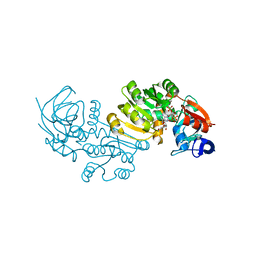 | | Crystal Structure of human P53 inducible oxidoreductase (TP53I3,PIG3) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, QUINONE OXIDOREDUCTASE | | Authors: | Pike, A.C.W, Shafqat, N, Debreczeni, J, Johansson, C, Haroniti, A, Gileadi, O, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, von Delft, F, Porte, S, Fita, I, Pares, J, Pares, X, Oppermann, U. | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-Dimensional Structure and Enzymatic Function of Proapoptotic Human P53-Inducible Quinone Oxidoreductase Pig3.
J.Biol.Chem., 284, 2009
|
|
2IQF
 
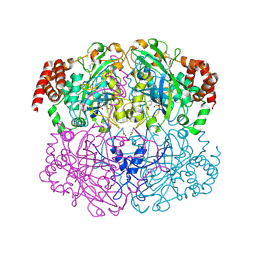 | | Crystal structure of Helicobacter pylori catalase compound I | | Descriptor: | ACETATE ION, Catalase, OXYGEN ATOM, ... | | Authors: | Loewen, P.C, Carpena, X, Fita, I. | | Deposit date: | 2006-10-13 | | Release date: | 2007-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structures and electronic configuration of compound I intermediates of Helicobacter pylori and Penicillium vitale catalases determined by X-ray crystallography and QM/MM density functional theory calculations.
J.Am.Chem.Soc., 129, 2007
|
|
2JJ4
 
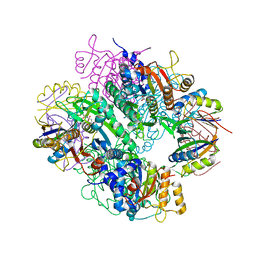 | | The complex of PII and acetylglutamate kinase from Synechococcus elongatus PCC7942 | | Descriptor: | ACETYLGLUTAMATE KINASE, N-ACETYL-L-GLUTAMATE, NITROGEN REGULATORY PROTEIN P-II | | Authors: | Llacer, J.L, Marco-Marin, C, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2007-07-04 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | The Crystal Structure of the Complex of Pii and Acetylglutamate Kinase Reveals How Pii Controls the Storage of Nitrogen as Arginine
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2OBY
 
 | | Crystal structure of Human P53 inducible oxidoreductase (TP53I3,PIG3) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative quinone oxidoreductase | | Authors: | Porte, S, Valencia, E, Farres, J, Fita, I, Pike, A.C.W, Shafqat, N, Debreczeni, J, Johansson, C, Haroniti, A, Gileadi, O, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, von Delft, F, Oppermann, U, Pares, X. | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human P53 inducible oxidoreductase (TP53I3, PIG3)
To be Published
|
|
5OBU
 
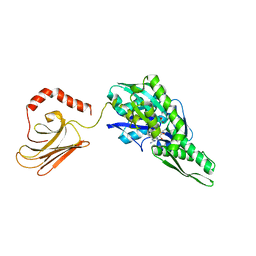 | | Mycoplasma genitalium DnaK deletion mutant lacking SBDalpha in complex with AMPPNP. | | Descriptor: | Chaperone protein DnaK, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
5OBY
 
 | | Mycoplasma genitalium DnaK-NBD in complex with AMP-PNP | | Descriptor: | Chaperone protein DnaK, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
5OBV
 
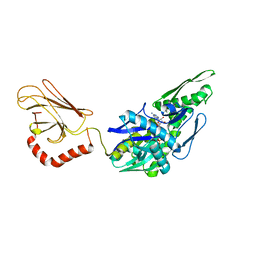 | | Mycoplasma genitalium DnaK deletion mutant lacking SBDalpha in complex with ADP and Pi. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein DnaK, PHOSPHATE ION | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
5OX7
 
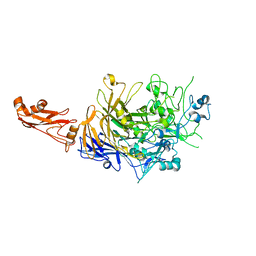 | |
5OBW
 
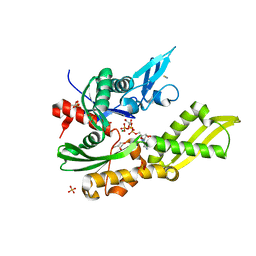 | | Mycoplasma genitalium DnaK-NBD in complex with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
5OBX
 
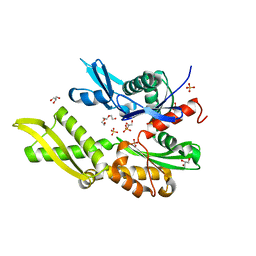 | | Mycoplasma genitalium DnaK-NBD | | Descriptor: | Chaperone protein DnaK, GLYCEROL, SULFATE ION, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
1GS5
 
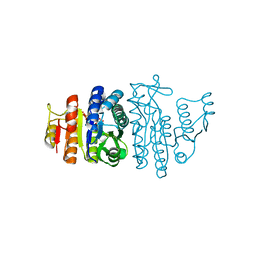 | | N-acetyl-L-glutamate kinase from Escherichia coli complexed with its substrate N-acetylglutamate and its substrate analog AMPPNP | | Descriptor: | ACETYLGLUTAMATE KINASE, MAGNESIUM ION, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2001-12-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Acetylglutamate Kinase, a Key Enzyme for Arginine Biosynthesis and a Prototype for the Amino Acid Kinase Enzyme Family, During Catalysis
Structure, 10, 2002
|
|
1GSJ
 
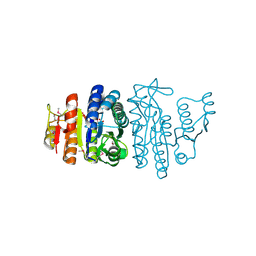 | | Selenomethionine substituted N-acetyl-L-glutamate kinase from Escherichia coli complexed with its substrate n-acetyl-L-glutamate and its substrate analog AMPPNP | | Descriptor: | ACETYLGLUTAMATE KINASE, MAGNESIUM ION, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2002-01-07 | | Release date: | 2002-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Acetylglutamate Kinase, a Key Enzyme for Arginine Biosynthesis and a Prototype for the Amino Acid Kinase Enzyme Family, During Catalysis
Structure, 10, 2002
|
|
1GMI
 
 | | Structure of the c2 domain from novel protein kinase C epsilon | | Descriptor: | MAGNESIUM ION, PROTEIN KINASE C, EPSILON TYPE | | Authors: | Ochoa, W.F, Garcia-Garcia, J, Fita, I, Corbalan-Garcia, S, Verdaguer, N, Gomez-Fernandez, J.C. | | Deposit date: | 2001-09-14 | | Release date: | 2001-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the C2 Domain from Novel Protein Kinase Cepsilon. A Membrane Binding Model for Ca(2+ )-Independent C2 Domains
J.Mol.Biol., 311, 2001
|
|
1MQT
 
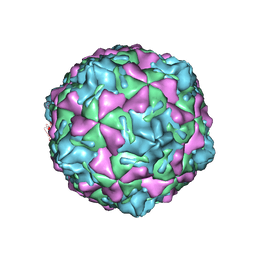 | | Swine Vesicular Disease Virus coat protein | | Descriptor: | OCTANOIC ACID (2-HYDROXY-1-HYDROXYMETHYL-HEPTADEC-3-ENYL)-AMIDE, Polyprotein, Polyprotein Capsid Protein | | Authors: | Verdaguer, N, Jimenez-Clavero, M.A, Fita, I, Ley, V. | | Deposit date: | 2002-09-17 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | STRUCTURE OF SWINE VESICULAR DISEASE VIRUS: MAPPING OF CHANGES OCCURRING DURING ADAPTATION OF HUMAN COXSACKIE B5 VIRUS TO INFECT SWINE
J.Virol., 77, 2003
|
|
1E19
 
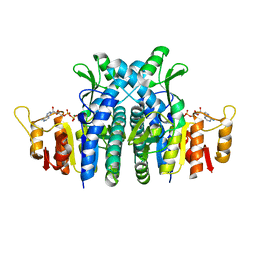 | | Structure of the carbamate kinase-like carbamoyl phosphate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMATE KINASE, MAGNESIUM ION | | Authors: | Ramon-Maiques, S, Marina, A, Uriarte, M, Fita, I, Rubio, V. | | Deposit date: | 2000-04-28 | | Release date: | 2000-07-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5-A Resolution Crystal Structure of the Carbamate Kinase-Like Carbamoyl Phosphate Synthetase from the Hyperthermophilic Archaeon Pyrococcus Furiosus, Bound to Adp, Confirms that This Thermoestable Enzyme is a Carbamate Kinase, and Provides Insights Into Substrate Binding and Stability in Carbamate Kinases
J.Mol.Biol., 299, 2000
|
|
1FPN
 
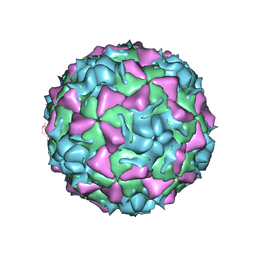 | | HUMAN RHINOVIRUS SEROTYPE 2 (HRV2) | | Descriptor: | COAT PROTEIN VP1, COAT PROTEIN VP2, COAT PROTEIN VP3, ... | | Authors: | Verdaguer, N, Blaas, D, Fita, I. | | Deposit date: | 2000-08-31 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human rhinovirus serotype 2 (HRV2).
J.Mol.Biol., 300, 2000
|
|
1DSY
 
 | | C2 DOMAIN FROM PROTEIN KINASE C (ALPHA) COMPLEXED WITH CA2+ AND PHOSPHATIDYLSERINE | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Verdaguer, N, Corbalan-Garcia, S, Ochoa, W.F, Fita, I, Gomez-Fernandez, J.C. | | Deposit date: | 2000-01-10 | | Release date: | 2000-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine.
EMBO J., 18, 1999
|
|
1EJO
 
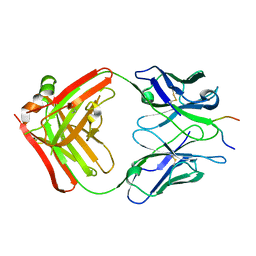 | | FAB FRAGMENT OF NEUTRALISING MONOCLONAL ANTIBODY 4C4 COMPLEXED WITH G-H LOOP FROM FMDV. | | Descriptor: | FMDV PEPTIDE, IGG2A MONOCLONAL ANTIBODY (HEAVY CHAIN), IGG2A MONOCLONAL ANTIBODY (LIGHT CHAIN) | | Authors: | Ochoa, W.F, Kalko, S.G, Gomes, P, Fita, I, Verdaguer, N. | | Deposit date: | 2000-03-03 | | Release date: | 2000-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A multiply substituted G-H loop from foot-and-mouth disease virus in complex with a neutralizing antibody: a role for water molecules.
J.Gen.Virol., 81, 2000
|
|
1P81
 
 | | Crystal structure of the D181E variant of catalase HPII from E. coli | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
1OHA
 
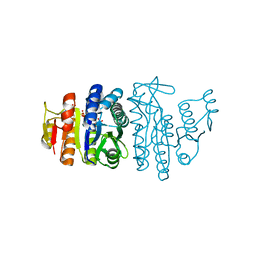 | | Acetylglutamate kinase from Escherichia coli complexed with MgADP and N-acetyl-L-glutamate | | Descriptor: | ACETATE ION, ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
