6T69
 
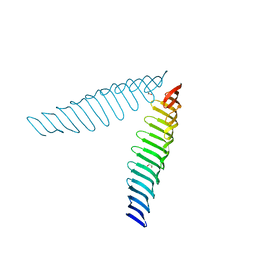 | | Crystal structure of Toxoplasma gondii Morn1(V shape) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Membrane occupation and recognition nexus protein MORN1, ... | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6T4D
 
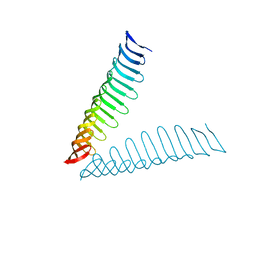 | | Crystal structure of Plasmodium falciparum Morn1 | | Descriptor: | Morn1, ZINC ION | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6T6Q
 
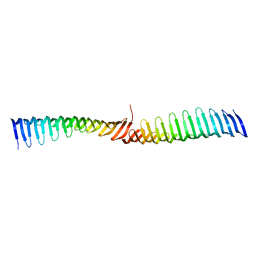 | | Crystal structure of Toxoplasma gondii Morn1 (extended conformation). | | Descriptor: | Membrane occupation and recognition nexus protein MORN1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6T68
 
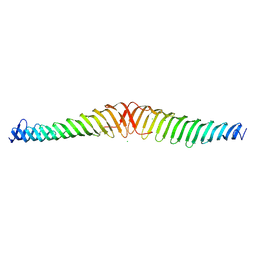 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | CHLORIDE ION, MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6T4R
 
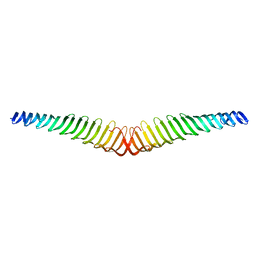 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6SL7
 
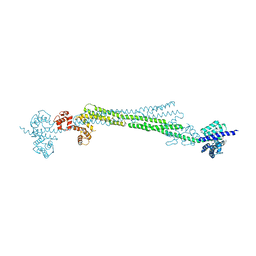 | |
6SL2
 
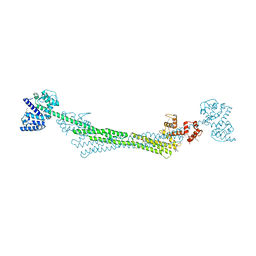 | | ALPHA-ACTININ FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Calponin homology domain protein putative, ... | | Authors: | Pinotsis, N, Khan, M.B, Djinovic-Carugo, K. | | Deposit date: | 2019-08-18 | | Release date: | 2020-08-26 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Calcium modulates the domain flexibility and function of an alpha-actinin similar to the ancestral alpha-actinin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SL3
 
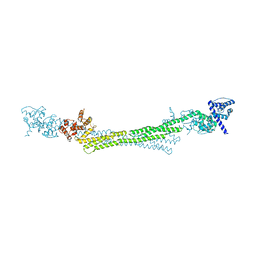 | |
5JWG
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with dipeptide Arg-Asp | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, ASPARTIC ACID, ... | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-12 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JXF
 
 | | Crystal structure of Flavobacterium psychrophilum DPP11 in complex with dipeptide Arg-Asp | | Descriptor: | ARGININE, ASPARTIC ACID, Asp/Glu-specific dipeptidyl-peptidase, ... | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K8Z
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 8.5) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Chlorite dismutase, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5JY0
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with substrate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Asp/Glu-specific dipeptidyl-peptidase, LEU-ASP-VAL | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JWF
 
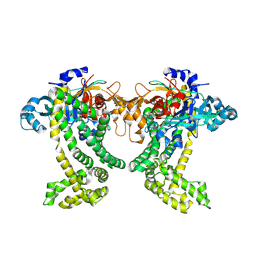 | | Crystal structure of Porphyromonas gingivalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-12 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JXP
 
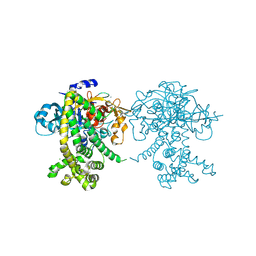 | | Crystal structure of Porphyromonas endodontalis DPP11 in alternate conformation | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CALCIUM ION, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K90
 
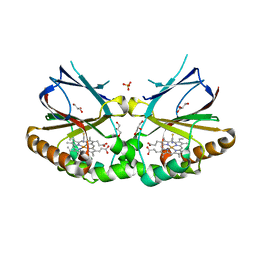 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with isothiocyanate | | Descriptor: | Chlorite dismutase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5JWI
 
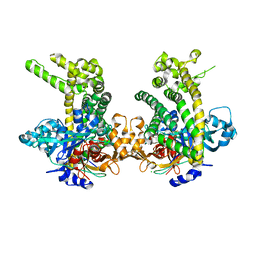 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with dipeptide Arg-Glu | | Descriptor: | ARGININE, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, ... | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-12 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K91
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with fluoride | | Descriptor: | Chlorite dismutase, FLUORIDE ION, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5JXK
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
6RWV
 
 | | Structure of apo-LmCpfC | | Descriptor: | Ferrochelatase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6386379 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
6RHW
 
 | | Crystal structure of human CD11b I-domain (CD11b-I) in complex with Staphylococcus aureus octameric bi-component leukocidin LukGH | | Descriptor: | Beta-channel forming cytolysin, DIMETHYL SULFOXIDE, Integrin alpha-M, ... | | Authors: | Trstenjak, N, Milic, D, Djinovic-Carugo, K, Badarau, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SV3
 
 | | Structure of coproheme-LmCpfC | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Ferrochelatase, GLYCEROL | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-09-17 | | Release date: | 2019-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64000869 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
6RHV
 
 | | Crystal structure of mouse CD11b I-domain (CD11b-I) in complex with Staphylococcus aureus octameric bi-component leukocidin LukGH (LukH K319A mutant) | | Descriptor: | Beta-channel forming cytolysin, DIMETHYL SULFOXIDE, Integrin alpha-M, ... | | Authors: | Trstenjak, N, Milic, D, Djinovic-Carugo, K, Badarau, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
9FEK
 
 | |
5EZU
 
 | | Crystal structure of the N-terminal domain of vaccinia virus immunomodulator A46 in complex with myristic acid. | | Descriptor: | MYRISTIC ACID, Protein A46 | | Authors: | Fedosyuk, S, Bezerra, G.A, Sammito, M, Uson, I, Skern, T. | | Deposit date: | 2015-11-26 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Vaccinia Virus Immunomodulator A46: A Lipid and Protein-Binding Scaffold for Sequestering Host TIR-Domain Proteins.
PLoS Pathog., 12, 2016
|
|
4WV8
 
 | | Crystal structure of a recombinant Vatairea macrocarpa seed lectin complexed with lactose | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Seed lectin, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Cunha, R.M.S, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-11-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural characterization of a Vatairea macrocarpa lectin in complex with a tumor-associated antigen: A new tool for cancer research.
Int.J.Biochem.Cell Biol., 72, 2016
|
|
