4I8D
 
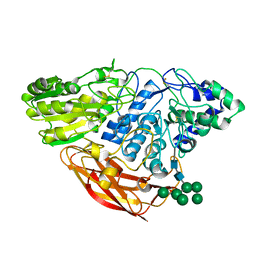 | | Crystal Structure of Beta-D-glucoside glucohydrolase from Trichoderma reesei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucoside glucohydrolase, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Helmich, K.E, Banerjee, G, Bianchetti, C.M, Gudmundsson, M, Sandgren, M, Walton, J.D, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Biochemical Characterization and Crystal Structures of a Fungal Family 3 beta-Glucosidase, Cel3A from Hypocrea jecorina.
J.Biol.Chem., 289, 2014
|
|
2RGZ
 
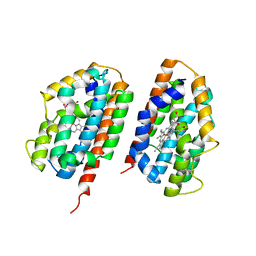 | | Ensemble refinement of the protein crystal structure of human heme oxygenase-2 C127A (HO-2) with bound heme | | Descriptor: | Heme oxygenase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bianchetti, C.M, Bingman, C.A, Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-10-05 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparison of Apo- and Heme-bound Crystal Structures of a Truncated Human Heme Oxygenase-2.
J.Biol.Chem., 282, 2007
|
|
3D89
 
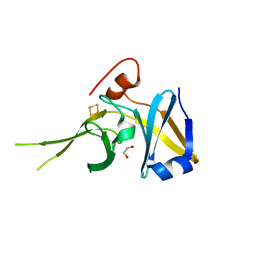 | | Crystal Structure of a Soluble Rieske Ferredoxin from Mus musculus | | Descriptor: | 1,2-ETHANEDIOL, FE2/S2 (INORGANIC) CLUSTER, Rieske domain-containing protein | | Authors: | Levin, E.J, McCoy, J.G, Elsen, N.L, Seder, K.D, Bingman, C.A, Wesenberg, G.E, Fox, B.G, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-05-22 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | X-ray structure of a soluble Rieske-type ferredoxin from Mus musculus.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3LHR
 
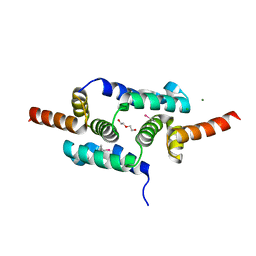 | | Crystal structure of the SCAN domain from Human ZNF24 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ETHYL MERCURY ION, ... | | Authors: | Volkman, B.F, Peterson, F.C, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SCAN domain from Human ZNF24
To be Published
|
|
3C9Q
 
 | | Crystal structure of the uncharacterized human protein C8orf32 with bound peptide | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, SULFATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the uncharacterized human protein C8orf32 with bound peptide.
To be Published
|
|
1JXC
 
 | | Minimized NMR structure of ATT, an Arabidopsis trypsin/chymotrypsin inhibitor | | Descriptor: | Putative trypsin inhibitor ATTI-2 | | Authors: | Zhao, Q, Chae, Y.K, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2001-09-06 | | Release date: | 2003-01-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of ATTp, an Arabidopsis thaliana Trypsin Inhibitor
Biochemistry, 41, 2002
|
|
3GAN
 
 | | Crystal structure of gene product from Arabidopsis thaliana At3g22680 with bound suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, CHLORIDE ION, Uncharacterized protein At3g22680 | | Authors: | Burgie, E.S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-02-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of gene product from Arabidopsis thaliana At3g22680 with bound suramin
To be Published
|
|
1VM0
 
 | | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT2G34160 | | Descriptor: | NITRATE ION, POTASSIUM ION, unknown protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-08-24 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT2G34160
To be published
|
|
1VKP
 
 | | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT5G08170, AGMATINE IMINOHYDROLASE | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, AGMATINE IMINOHYDROLASE, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g08170
To be published
|
|
3H7C
 
 | | Crystal Structure of Arabidopsis thaliana Agmatine Deiminase from Cell Free Expression | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, Agmatine deiminase, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Arabidopsis thaliana Agmatine Deiminase
To be Published
|
|
3PKQ
 
 | | Q83D Variant of S. Enterica RmlA with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Chang, A, Moretti, R, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution.
J.Biol.Chem., 286, 2011
|
|
3PKP
 
 | | Q83S Variant of S. Enterica RmlA with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Glucose-1-phosphate thymidylyltransferase, MAGNESIUM ION | | Authors: | Chang, A, Moretti, R, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expanding the Nucleotide and Sugar 1-Phosphate Promiscuity of Nucleotidyltransferase RmlA via Directed Evolution.
J.Biol.Chem., 286, 2011
|
|
3IHR
 
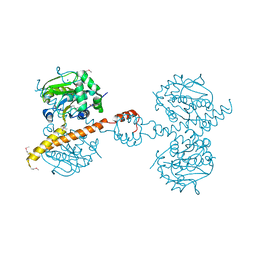 | | Crystal Structure of Uch37 | | Descriptor: | FORMIC ACID, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-11 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of human Uch37.
Proteins, 80, 2012
|
|
2BDU
 
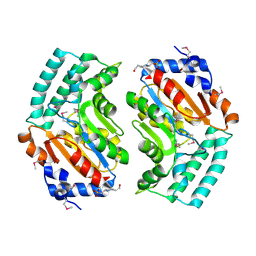 | | X-Ray Structure of a Cytosolic 5'-Nucleotidase III from Mus Musculus MM.158936 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
3RSC
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CalG2, Calicheamicin T0, PHOSPHATE ION, ... | | Authors: | Chang, A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-05-02 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3H7K
 
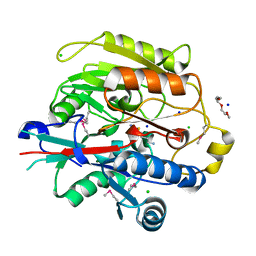 | |
2BAI
 
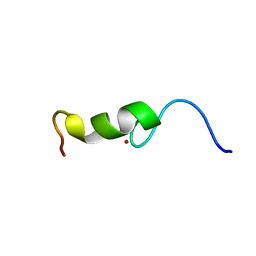 | | The Zinc finger domain of Mengovirus Leader polypeptide | | Descriptor: | Genome polyprotein, ZINC ION | | Authors: | Cornilescu, C.C, Porter, F.W, Qin, Z, Lee, M.S, Palmenberg, A.C, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the mengovirus Leader protein zinc-finger domain.
Febs Lett., 582, 2008
|
|
2EVN
 
 | | NMR solution structures of At1g77540 | | Descriptor: | Protein At1g77540 | | Authors: | Tyler, R.C, Singh, S, Tonelli, M, Min, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
3OTI
 
 | | Crystal Structure of CalG3, Calicheamicin Glycostyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CHLORIDE ION, CalG3, Calicheamicin T0, ... | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3BZB
 
 | | Crystal structure of uncharacterized protein CMQ451C from the primitive red alga Cyanidioschyzon merolae | | Descriptor: | Uncharacterized protein | | Authors: | McCoy, J.G, Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-01-17 | | Release date: | 2008-01-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of uncharacterized protein CMQ451C from the primitive red alga Cyanidioschyzon merolae.
To be Published
|
|
3DCY
 
 | |
3OTG
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form | | Descriptor: | CHLORIDE ION, CalG1, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1VK0
 
 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450 | | Descriptor: | hypothetical protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-12 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450
To be published
|
|
3IA8
 
 | |
3IAU
 
 | | The structure of the processed form of threonine deaminase isoform 2 from Solanum lycopersicum | | Descriptor: | ACETATE ION, POLYETHYLENE GLYCOL (N=34), SULFATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-28 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Adaptive evolution of threonine deaminase in plant defense against insect herbivores.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
