1CUI
 
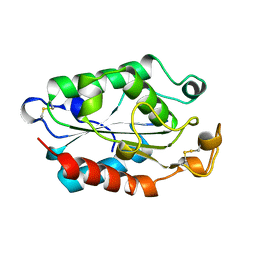 | | CUTINASE, S120A MUTANT | | Descriptor: | CUTINASE | | Authors: | Martinez, C, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1LCO
 
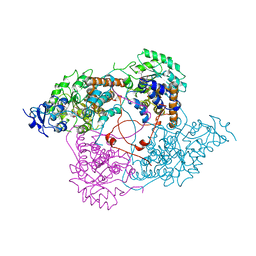 | |
1CUV
 
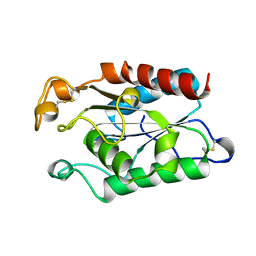 | | CUTINASE, A85F MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUJ
 
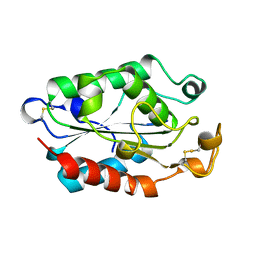 | | CUTINASE, S120C MUTANT | | Descriptor: | CUTINASE | | Authors: | Martinez, C, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUY
 
 | | CUTINASE, L189F MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUZ
 
 | | CUTINASE, L81G, L182G MUTANT | | Descriptor: | CUTINASE | | Authors: | Nicolas, A, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUW
 
 | | CUTINASE, G82A, A85F, V184I, A185L, L189F MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUE
 
 | | CUTINASE, Q121L MUTANT | | Descriptor: | CUTINASE | | Authors: | Martinez, C, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUC
 
 | |
1CUX
 
 | | CUTINASE, L114Y MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUG
 
 | | CUTINASE, R17E, N172K MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUU
 
 | | CUTINASE, A199C MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUB
 
 | |
1CUD
 
 | |
1CUF
 
 | | CUTINASE, R156L MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUH
 
 | | CUTINASE, R196E MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUA
 
 | | CUTINASE, N172K MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
3BJH
 
 | | Soft-SAD crystal structure of a pheromone binding protein from the honeybee Apis mellifera L. | | Descriptor: | GLYCEROL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Lartigue, A, Gruez, A, Briand, L, Blon, F, Bezirard, V, Walsh, M, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sulfur single-wavelength anomalous diffraction crystal structure of a pheromone-binding protein from the honeybee Apis mellifera L.
J.Biol.Chem., 279, 2004
|
|
2H36
 
 | |
4O9H
 
 | | Structure of Interleukin-6 in complex with a Camelid Fab fragment | | Descriptor: | Heavy Chain of the Camelid Fab fragment 61H7, Interleukin-6, Light Chain of the Camelid Fab fragment 61H7 | | Authors: | Klarenbeek, A, Blanchetot, C, Schragel, G, Sadi, A.S, Ongenae, N, Hemrika, W, Wijdenes, J, Spinelli, S, Desmyter, A, Cambillau, C, Hultberg, A, Kretz-rommel, A, Dreier, T, De haard, H.J.W, Roovers, R.C. | | Deposit date: | 2014-01-02 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Combining residues of naturally-occurring Camelid somatic affinity variants yields ultra-potent human therapeutic IL-6 antibodies
To be Published
|
|
2WZP
 
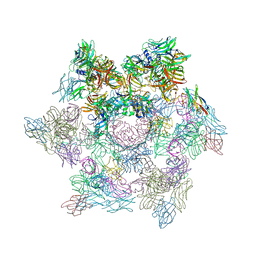 | | Structures of Lactococcal Phage p2 Baseplate Shed Light on a Novel Mechanism of Host Attachment and Activation in Siphoviridae | | Descriptor: | CAMELID VHH5, LACTOCOCCAL PHAGE P2 ORF15, LACTOCOCCAL PHAGE P2 ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2009-12-01 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1CUS
 
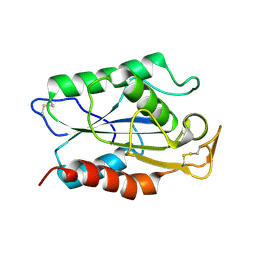 | |
5E7F
 
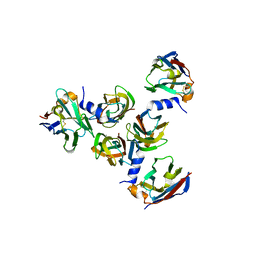 | | Complex between lactococcal phage Tuc2009 RBP head domain and a nanobody (L06) | | Descriptor: | Major structural protein 1, nanobody L06 | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-12 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
5E7B
 
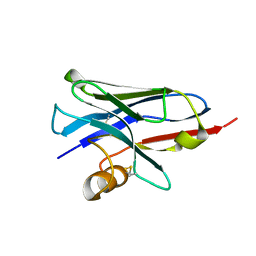 | | Structure of a nanobody (vHH) from camel against phage Tuc2009 RBP (BppL, ORF53) | | Descriptor: | nanobody nano-L06 | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-12 | | Release date: | 2015-12-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
5E7T
 
 | | Structure of the tripod (BppUct-A-L) from the baseplate of bacteriophage Tuc2009 | | Descriptor: | CALCIUM ION, Major structural protein 1, Minor structural protein 4, ... | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
