3RSX
 
 | |
3RTM
 
 | |
3RVI
 
 | |
3RU1
 
 | |
3RTH
 
 | |
3RTN
 
 | |
3C6L
 
 | |
3C60
 
 | |
3ZFJ
 
 | | N-terminal domain of pneumococcal PhtD protein with bound Zn(II) | | Descriptor: | PNEUMOCOCCAL HISTIDINE TRIAD PROTEIN D, ZINC ION | | Authors: | Bersch, B, Bougault, C, Favier, A, Gabel, F, Roux, L, Vernet, T, Durmort, C. | | Deposit date: | 2012-12-11 | | Release date: | 2013-11-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | New Insights Into Histidine Triad Proteins: Solution Structure of a Streptococcus Pneumoniae Phtd Domain and Zinc Transfer to Adcaii.
Plos One, 8, 2013
|
|
5QTZ
 
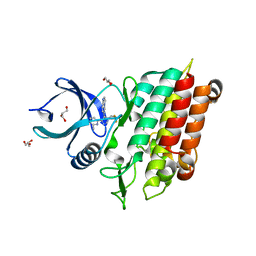 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH 6-[1-(2,2-DIFLUOROETHYL)-4-(6-METHYLPYRIDIN-2-YL)-1H-IMIDAZOL-5-YL]IMIDAZO[1,2-A]PYRIDINE | | Descriptor: | 6-[1-(2,2-difluoroethyl)-4-(6-methylpyridin-2-yl)-1H-imidazol-5-yl]imidazo[1,2-a]pyridine, GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of BMS-986260, a Potent, Selective, and Orally Bioavailable TGF beta R1 Inhibitor as an Immuno-oncology Agent.
Acs Med.Chem.Lett., 11, 2020
|
|
5QU0
 
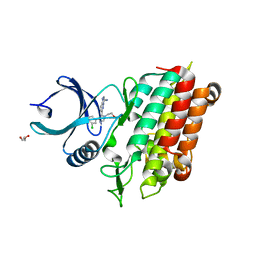 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH 6-[4-(3-CHLORO-4-FLUOROPHENYL)-1-(2-HYDROXYETHYL)-1H-IMIDAZOL-5-YL]IMIDAZO[1,2-B]PYRIDAZINE-3-CARBONITRILE | | Descriptor: | 6-[4-(3-chloro-4-fluorophenyl)-1-(2-hydroxyethyl)-1H-imidazol-5-yl]imidazo[1,2-b]pyridazine-3-carbonitrile, GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of BMS-986260, a Potent, Selective, and Orally Bioavailable TGF beta R1 Inhibitor as an Immuno-oncology Agent.
Acs Med.Chem.Lett., 11, 2020
|
|
5TR4
 
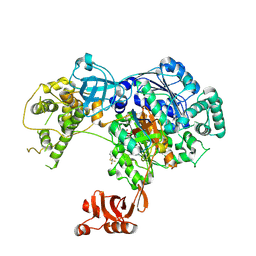 | | Structure of Ubiquitin activating enzyme (Uba1) in complex with ubiquitin and TAK-243 | | Descriptor: | Ubiquitin, Ubiquitin-activating enzyme E1 1, [(1~{R},2~{R},3~{S},4~{R})-2,3-bis(oxidanyl)-4-[[2-[3-(trifluoromethylsulfanyl)phenyl]pyrazolo[1,5-a]pyrimidin-7-yl]amino]cyclopentyl]methyl sulfamate | | Authors: | Sintchak, M.D. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification and Characterization of a Small Molecule Inhibitor of the Ubiquitin Activating Enzyme (TAK-243)
To be published
|
|
