6YN1
 
 | | Crystal structure of histone chaperone APLF acidic domain bound to the histone H2A-H2B-H3-H4 octamer | | Descriptor: | Aprataxin and PNK-like factor, CHLORIDE ION, GLYCEROL, ... | | Authors: | Corbeski, I, Guo, X, Van Ingen, H, Sixma, T.K. | | Deposit date: | 2020-04-10 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Sci Adv, 8, 2022
|
|
4OC7
 
 | | Retinoic acid receptor alpha in complex with (E)-3-(3'-allyl-6-hydroxy-[1,1'-biphenyl]-3-yl)acrylic acid and a fragment of the coactivator TIF2 | | Descriptor: | (2E)-3-[6-hydroxy-3'-(prop-2-en-1-yl)biphenyl-3-yl]prop-2-enoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Leysen, S, Scheepstra, M, Brunsveld, L, Milroy, L.G, Ottmann, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A natural-product switch for a dynamic protein interface.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1GDC
 
 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | Deposit date: | 1994-03-15 | | Release date: | 1994-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
1EFA
 
 | |
2GDA
 
 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | Deposit date: | 1994-03-15 | | Release date: | 1994-06-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
3QZU
 
 | | Crystal structure of Bacillus subtilis Lipase A 7-fold mutant; the outcome of directed evolution towards thermostability | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipase estA, ... | | Authors: | Pijning, T, Augustyniak, W, Reetz, M.T, Dijkstra, B.W. | | Deposit date: | 2011-03-07 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biophysical characterization of mutants of Bacillus subtilis lipase evolved for thermostability: Factors contributing to increased activity retention.
Protein Sci., 21, 2012
|
|
1D1N
 
 | | SOLUTION STRUCTURE OF THE FMET-TRNAFMET BINDING DOMAIN OF BECILLUS STEAROTHERMOPHILLUS TRANSLATION INITIATION FACTOR IF2 | | Descriptor: | INITIATION FACTOR 2 | | Authors: | Meunier, S, Spurio, S, Czisch, M, Wechselberger, R, Gueunneugues, M. | | Deposit date: | 1999-09-20 | | Release date: | 2000-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the fMet-tRNA(fMet)-binding domain of B. stearothermophilus initiation factor IF2.
EMBO J., 19, 2000
|
|
3PAT
 
 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Padilla, A, Cave, A, Parello, J, Etienne, G, Baldellon, C. | | Deposit date: | 1994-03-22 | | Release date: | 1994-07-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin
To be Published
|
|
2YB6
 
 | | Native human Rad6 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, UBIQUITIN-CONJUGATING ENZYME E2 B | | Authors: | Hibbert, R.G, Sixma, T.K. | | Deposit date: | 2011-03-02 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | E3 Ligase Rad18 Promotes Monoubiquitination Rather Than Ubiquitin Chain Formation by E2 Enzyme Rad6.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2YBF
 
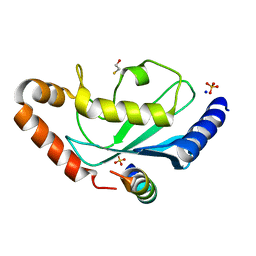 | | Complex of Rad18 (Rad6 binding domain) with Rad6b | | Descriptor: | BETA-MERCAPTOETHANOL, E3 UBIQUITIN-PROTEIN LIGASE RAD18, SODIUM ION, ... | | Authors: | Hibbert, R.G, Sixma, T.K. | | Deposit date: | 2011-03-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | E3 Ligase Rad18 Promotes Monoubiquitination Rather Than Ubiquitin Chain Formation by E2 Enzyme Rad6.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
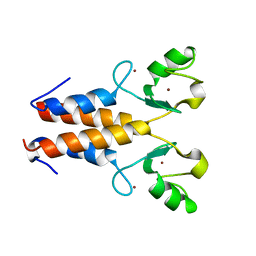
2Y43
 
 | |
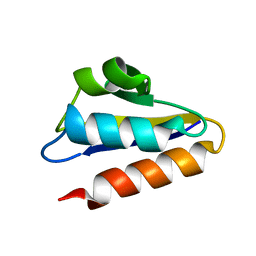
1PFH
 
 | |
2GVS
 
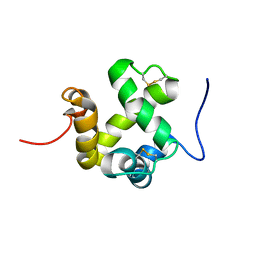 | | NMR solution structure of CSPsg4 | | Descriptor: | chemosensory protein CSP-sg4 | | Authors: | Tomaselli, S, Crescenzi, O, Sanfelice, D, Ab, E, Tancredi, T, Picone, D. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Chemosensory Protein from the Desert Locust Schistocerca gregaria(,).
Biochemistry, 45, 2006
|
|
1WCO
 
 | | The solution structure of the nisin-lipid II complex | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALA-FGA-LYS-DAL-DAL PEPTIDE, ... | | Authors: | Hsu, S.-T.D, Breukink, E, Tischenko, E, Lutters, M.A.G, de Kruijff, B, Kaptein, R, Bonvin, A.M.J.J, van Nuland, N.A.J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-03-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics.
Nat. Struct. Mol. Biol., 11, 2004
|
|
2LFJ
 
 | |
1D7E
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 1999-10-17 | | Release date: | 2000-03-31 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Conformational substates in different crystal forms of the photoactive yellow protein--correlation with theoretical and experimental flexibility.
Protein Sci., 9, 2000
|
|
2PAS
 
 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Padilla, A, Cave, A, Parello, J, Etienne, G, Baldellon, C. | | Deposit date: | 1994-03-22 | | Release date: | 1994-06-22 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin
To be Published
|
|
