4GQY
 
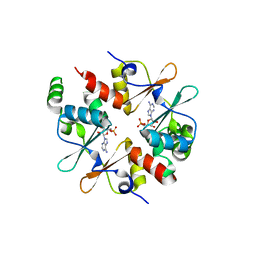 | | Crystal structure of CBSX2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CBS domain-containing protein CBSX2, chloroplastic | | Authors: | Jeong, B.C, Song, H.K. | | Deposit date: | 2012-08-24 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Change in single cystathionine beta-synthase domain-containing protein from a bent to flat conformation upon adenosine monophosphate binding
J.Struct.Biol., 183, 2013
|
|
4HNZ
 
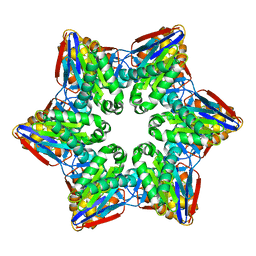 | |
4HO7
 
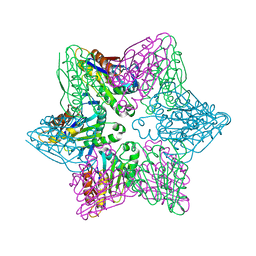 | |
2AEH
 
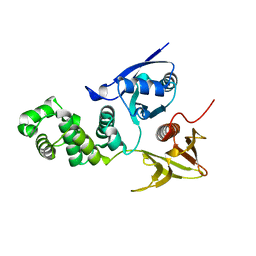 | | Focal adhesion kinase 1 | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Ceccarelli, D.F, Song, H.K, Poy, F, Schaller, M.D, Eck, M.J. | | Deposit date: | 2005-07-22 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal Structure of the FERM Domain of Focal Adhesion Kinase
J.Biol.Chem., 281, 2006
|
|
3PO0
 
 | | Crystal structure of SAMP1 from Haloferax volcanii | | Descriptor: | ACETATE ION, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Jeong, Y.J, Jeong, B.-C, Song, H.K. | | Deposit date: | 2010-11-21 | | Release date: | 2011-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of ubiquitin-like small archaeal modifier protein 1 (SAMP1) from Haloferax volcanii.
Biochem.Biophys.Res.Commun., 405, 2011
|
|
3RUI
 
 | | Crystal structure of Atg7C-Atg8 complex | | Descriptor: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Hong, S.B, Kim, B.W, Song, H.K. | | Deposit date: | 2011-05-05 | | Release date: | 2011-11-23 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SL7
 
 | | Crystal structure of CBS-pair protein, CBSX2 from Arabidopsis thaliana | | Descriptor: | ACETATE ION, CBS domain-containing protein CBSX2, GLYCEROL | | Authors: | Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2011-06-24 | | Release date: | 2011-11-09 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Single cystathionine beta-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis
Plant Cell, 23, 2011
|
|
3RUJ
 
 | |
2DS6
 
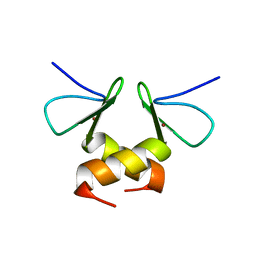 | | Structure of the ZBD in the tetragonal crystal form | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2AL6
 
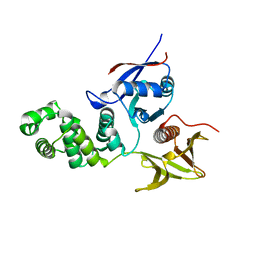 | | FERM domain of Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Ceccarelli, D.F, Song, H.K, Poy, F, Schaller, M.D, Eck, M.J. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the FERM Domain of Focal Adhesion Kinase
J.Biol.Chem., 281, 2006
|
|
2DS8
 
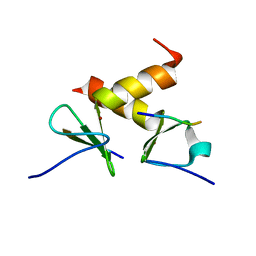 | | Structure of the ZBD-XB complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, SspB-tail peptide, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Kim, H.W, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
1M27
 
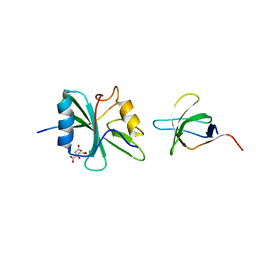 | | Crystal structure of SAP/FynSH3/SLAM ternary complex | | Descriptor: | CITRATE ANION, Proto-oncogene tyrosine-protein kinase FYN, SH2 domain protein 1A, ... | | Authors: | Chan, B, Griesbach, J, Song, H.K, Poy, F, Terhorst, C, Eck, M.J. | | Deposit date: | 2002-06-21 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SAP couples Fyn to SLAM immune receptors.
NAT.CELL BIOL., 5, 2003
|
|
4TQ1
 
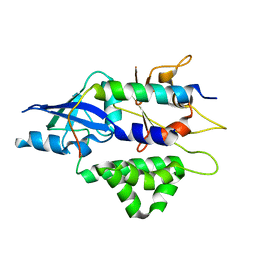 | | Crystal structure of human ATG5-TECAIR | | Descriptor: | Autophagy protein 5, Tectonin beta-propeller repeat-containing protein 1 | | Authors: | Kim, J.H, Hong, S.B, Song, H.K. | | Deposit date: | 2014-06-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners
Autophagy, 11, 2015
|
|
4TQ0
 
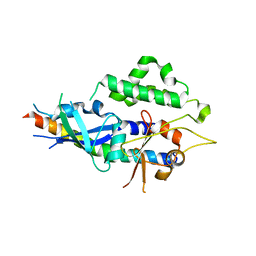 | | Crystal structure of human ATG5-ATG16N69 | | Descriptor: | Autophagy protein 5, Autophagy-related protein 16-1 | | Authors: | Kim, J.H, Hong, S.B, Song, H.K. | | Deposit date: | 2014-06-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners
Autophagy, 11, 2015
|
|
2DS7
 
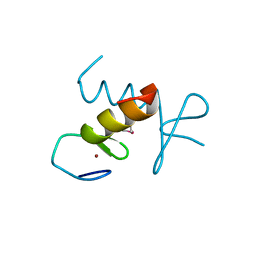 | | Structure of the ZBD in the hexagonal crystal form | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
3NIL
 
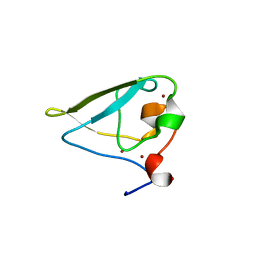 | | The structure of UBR box (RDAA) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NII
 
 | | The structure of UBR box (KIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide KIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIM
 
 | | The structure of UBR box (RRAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RRAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIJ
 
 | | The structure of UBR box (HIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide HIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIN
 
 | | The structure of UBR box (RLGES) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RLGES, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIT
 
 | | The structure of UBR box (native1) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIH
 
 | | The structure of UBR box (RIAAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIK
 
 | | The structure of UBR box (REAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIS
 
 | | The structure of UBR box (native2) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
2H8G
 
 | | 5'-Methylthioadenosine Nucleosidase from Arabidopsis thaliana | | Descriptor: | 5'-Methylthioadenosine Nucleosidase, ADENINE | | Authors: | Park, E.Y, Oh, S.I, Nam, M.J, Shin, J.S, Kim, K.N, Song, H.K. | | Deposit date: | 2006-06-07 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine nucleosidase from Arabidopsis thaliana at 1.5-A resolution
Proteins, 65, 2006
|
|
