8DDH
 
 | |
8DDF
 
 | | Quasi-racemic mixture of L-FWF and D-FYF peptide reveals rippled beta-sheet | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, DPN-DTY-DPN, PHE-TRP-PHE | | Authors: | Sawaya, M.R, Hazari, A, Eisenberg, D.E. | | Deposit date: | 2022-06-18 | | Release date: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The rippled beta-sheet layer configuration-a novel supramolecular architecture based on predictions by Pauling and Corey.
Chem Sci, 13, 2022
|
|
1F1C
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C549 | | Descriptor: | CYTOCHROME C549, HEME C | | Authors: | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O, Krogmann, D.W. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
1F1F
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM ARTHROSPIRA MAXIMA | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Serag, A.A, Sawaya, M.R, Krogmann, D.W, Yeates, T.O. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
5K2H
 
 | |
5K2F
 
 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | Descriptor: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2G
 
 | |
8T84
 
 | | Racemic mixture of amyloid beta segment 35-MVGGVV-40 forms heterochiral rippled beta-sheet, includes hexafluoroisopropanol | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, Racemic mixture of amyloid beta segment 35-MVGGVV-40 | | Authors: | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Racemic Peptides from Amyloid beta and Amylin Form Rippled beta-Sheets Rather Than Pleated beta-Sheets.
J.Am.Chem.Soc., 145, 2023
|
|
8T86
 
 | |
8T82
 
 | | Racemic mixture of amyloid beta segment 35-MVGGVV-40 forms heterochiral rippled beta-sheet, includes pentafluoropropionic acid | | Descriptor: | amyloid beta segment 35-MVGGVV-40, racemic mixture, pentafluoropropanoic acid | | Authors: | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Racemic Peptides from Amyloid beta and Amylin Form Rippled beta-Sheets Rather Than Pleated beta-Sheets.
J.Am.Chem.Soc., 145, 2023
|
|
8T89
 
 | |
2ON9
 
 | |
2OL9
 
 | |
2OMP
 
 | |
2OLX
 
 | |
2ONA
 
 | |
2ONV
 
 | |
2OMQ
 
 | |
2ONW
 
 | |
1MQ7
 
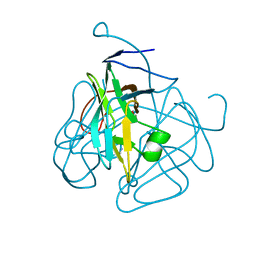 | | CRYSTAL STRUCTURE OF DUTPASE FROM MYCOBACTERIUM TUBERCULOSIS (RV2697C) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Sawaya, M.R, Chan, S, Segelke, B.W, Lekin, T, Heike, K, Cho, U.S, Naranjo, C, Perry, L.J, Yeates, T.O, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-09-13 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
3GGS
 
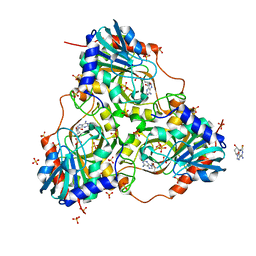 | | Human purine nucleoside phosphorylase double mutant E201Q,N243D complexed with 2-fluoro-2'-deoxyadenosine | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Sawaya, M.R, Afshar, S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of a mutant human purine nucleoside phosphorylase with the prodrug, 2-fluoro-2'-deoxyadenosine and the cytotoxic drug, 2-fluoroadenine.
Protein Sci., 18, 2009
|
|
3GB9
 
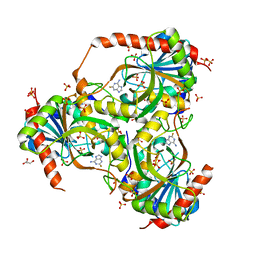 | | Human purine nucleoside phosphorylase double mutant E201Q,N243D complexed with 2-fluoroadenine | | Descriptor: | 2-fluoroadenine, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Sawaya, M.R, Afshar, S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a mutant human purine nucleoside phosphorylase with the prodrug, 2-fluoro-2'-deoxyadenosine and the cytotoxic drug, 2-fluoroadenine.
Protein Sci., 18, 2009
|
|
3NI3
 
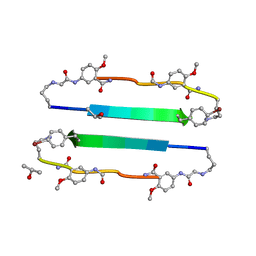 | | 54-Membered ring macrocyclic beta-sheet peptide | | Descriptor: | 54-membered ring macrocyclic beta-sheet peptide, ISOPROPYL ALCOHOL | | Authors: | Sawaya, M.R, Eisenberg, D, Nowick, J.S, Korman, T.P, Khakshoor, O. | | Deposit date: | 2010-06-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray crystallographic structure of an artificial beta-sheet dimer.
J.Am.Chem.Soc., 132, 2010
|
|
3HLC
 
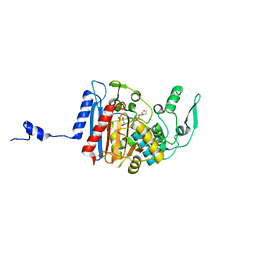 | | Simvastatin Synthase (LovD) from Aspergillus terreus, S5 mutant, unliganded | | Descriptor: | GLYCEROL, TETRAETHYLENE GLYCOL, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Laidman, J, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
