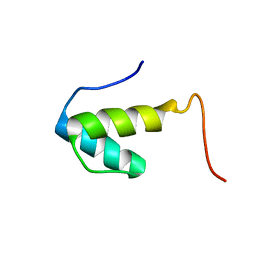
2UWQ
 
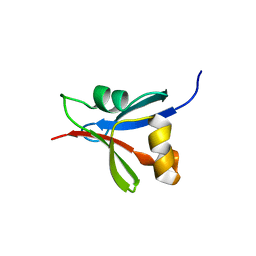 | |
2WXC
 
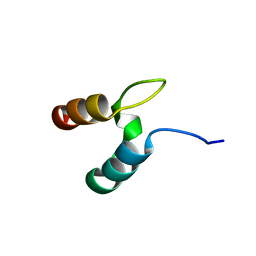 | | The folding mechanism of BBL: Plasticity of transition-state structure observed within an ultrafast folding protein family. | | Descriptor: | DIHYDROLIPOYLTRANSSUCCINASE | | Authors: | Neuweiler, H, Sharpe, T.D, Rutherford, T.J, Johnson, C.M, Allen, M.D, Ferguson, N, Fersht, A.R. | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Folding Mechanism of Bbl: Plasticity of Transition-State Structure Observed within an Ultrafast Folding Protein Family.
J.Mol.Biol., 390, 2009
|
|
2VY5
 
 | | U11-48K CHHC Zn-finger protein domain | | Descriptor: | U11/U12 SMALL NUCLEAR RIBONUCLEOPROTEIN 48 KDA PROTEIN, ZINC ION | | Authors: | Tidow, H, Andreeva, A, Rutherford, T.J, Fersht, A.R. | | Deposit date: | 2008-07-18 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U11-48K CHHC zinc-finger domain that specifically binds the 5' splice site of U12-type introns.
Structure, 17, 2009
|
|
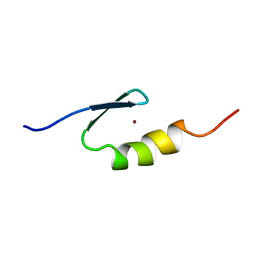
2WQG
 
 | | SAP domain from Tho1: L31W (fluorophore) mutant | | Descriptor: | PROTEIN THO1 | | Authors: | Dodson, C.A, Ferguson, N, Rutherford, T.J, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2009-08-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Engineering a Two-Helix Bundle Protein for Folding Studies.
Protein Eng.Des.Sel., 23, 2010
|
|
1BAO
 
 | |
1BSC
 
 | |
1BAN
 
 | |
1BSD
 
 | |
1BRG
 
 | |
1BSB
 
 | |
1BSE
 
 | |
1BSA
 
 | |
1BTB
 
 | |
1B2Z
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B20
 
 | | DELETION OF A BURIED SALT-BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1AB7
 
 | |
1COA
 
 | | THE EFFECT OF CAVITY CREATING MUTATIONS IN THE HYDROPHOBIC CORE OF CHYMOTRYPSIN INHIBITOR 2 | | Descriptor: | CHYMOTRYPSIN INHIBITOR 2 | | Authors: | Jackson, S.E, Moracci, M, Elmasry, N, Johnson, C.M, Fersht, A.R. | | Deposit date: | 1993-05-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effect of cavity-creating mutations in the hydrophobic core of chymotrypsin inhibitor 2.
Biochemistry, 32, 1993
|
|
1BNE
 
 | | BARNASE A43C/S80C DISULFIDE MUTANT | | Descriptor: | BARNASE | | Authors: | Clarke, J, Henrick, K, Fersht, A.R. | | Deposit date: | 1995-03-31 | | Release date: | 1995-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Disulfide mutants of barnase. I: Changes in stability and structure assessed by biophysical methods and X-ray crystallography.
J.Mol.Biol., 253, 1995
|
|
1B2X
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 7.5 FROM A CRYO_COOLED CRYSTAL AT 100K | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Harrison, P, Vaughan, C.K, Buckle, A.M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1BRN
 
 | |
1B21
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1BNG
 
 | | BARNASE S85C/H102C DISULFIDE MUTANT | | Descriptor: | BARNASE | | Authors: | Clarke, J, Henrick, K, Fersht, A.R. | | Deposit date: | 1995-03-31 | | Release date: | 1995-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Disulfide mutants of barnase. I: Changes in stability and structure assessed by biophysical methods and X-ray crystallography.
J.Mol.Biol., 253, 1995
|
|
1BTA
 
 | |
1BNI
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 6.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
1BNJ
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 9.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
