1FST
 
 | | CRYSTAL STRUCTURE OF TRUNCATED HUMAN RHOGDI TRIPLE MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1 | | Authors: | Longenecker, K.L, Garrard, S.M, Sheffield, P.J, Derewenda, Z.S. | | Deposit date: | 2000-09-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein crystallization by rational mutagenesis of surface residues: Lys to Ala mutations promote crystallization of RhoGDI.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HTJ
 
 | | STRUCTURE OF THE RGS-LIKE DOMAIN FROM PDZ-RHOGEF | | Descriptor: | KIAA0380 | | Authors: | Longenecker, K.L, Lewis, M.E, Chikumi, H, Gutkind, J.S, Derewenda, Z.S. | | Deposit date: | 2000-12-29 | | Release date: | 2001-07-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the RGS-like domain from PDZ-RhoGEF: linking heterotrimeric g protein-coupled signaling to Rho GTPases.
Structure, 9, 2001
|
|
1THT
 
 | | STRUCTURE OF A MYRISTOYL-ACP-SPECIFIC THIOESTERASE FROM VIBRIO HARVEYI | | Descriptor: | THIOESTERASE | | Authors: | Lawson, D.M, Derewenda, U, Serre, L, Ferri, S, Szitter, R, Wei, Y, Meighen, E.A, Derewenda, Z.S. | | Deposit date: | 1994-04-19 | | Release date: | 1995-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a myristoyl-ACP-specific thioesterase from Vibrio harveyi.
Biochemistry, 33, 1994
|
|
1UUJ
 
 | | N-terminal domain of Lissencephaly-1 protein (Lis-1) | | Descriptor: | ACETATE ION, BENZOIC ACID, PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB ALPHA SUBUNIT, ... | | Authors: | Cooper, D.R, Kim, M.H, Devedjiev, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2003-12-22 | | Release date: | 2004-07-29 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structure of the N-Terminal Domain of the Product of the Lissencephaly Gene Lis1 and its Functional Implications
Structure, 12, 2004
|
|
1V1T
 
 | | Crystal structure of the PDZ tandem of human syntenin in complex with TNEYKV peptide | | Descriptor: | BENZOIC ACID, SYNTENIN 1, TNEYKV PEPTIDE | | Authors: | Grembecka, J, Cierpicki, T, Devedjiev, Y, Cooper, D.R, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2004-04-23 | | Release date: | 2005-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Binding of the Pdz Tandem of Syntenin to Target Proteins.
Biochemistry, 45, 2006
|
|
1TBU
 
 | |
1SBR
 
 | | The structure and function of B. subtilis YkoF gene product: the complex with thiamin | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, CALCIUM ION, ykoF | | Authors: | Devedjiev, Y, Surendranath, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure and Ligand Binding Properties of the B.subtilis YkoF Gene Product, a Member of a Novel Family of Thiamin/HMP-binding Proteins
J.Mol.Biol., 343, 2004
|
|
1S99
 
 | | The structure and function of B. subtilis YkoF gene product: ligand free protein | | Descriptor: | ACETATE ION, CALCIUM ION, ykoF | | Authors: | Devedjiev, Y, Surendranath, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2004-02-04 | | Release date: | 2004-10-05 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Structure and Ligand Binding Properties of the B.subtilis YkoF Gene Product, a Member of a Novel Family of Thiamin/HMP-binding Proteins
J.Mol.Biol., 343, 2004
|
|
1ES9
 
 | | X-RAY CRYSTAL STRUCTURE OF R22K MUTANT OF THE MAMMALIAN BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASES (PAF-AH) | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB GAMMA SUBUNIT | | Authors: | McMullen, T.W.P, Li, J, Sheffield, P.J, Aoki, J, Martin, T.W, Arai, H, Inoue, K, Derewenda, Z.S. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The functional implications of the dimerization of the catalytic subunits of the mammalian brain platelet-activating factor acetylhydrolase (Ib).
Protein Eng., 13, 2000
|
|
1UXO
 
 | | The crystal structure of the ydeN gene product from B. subtilis | | Descriptor: | Putative hydrolase YdeN | | Authors: | Janda, I.K, Devedjiev, Y, Cooper, D.R, Chruszcz, M, Derewenda, U, Gabrys, A, Minor, W, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-27 | | Release date: | 2004-05-27 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Harvesting the high-hanging fruit: the structure of the YdeN gene product from Bacillus subtilis at 1.8 angstroms resolution.
Acta Crystallogr. D Biol. Crystallogr., 60, 2004
|
|
1VZY
 
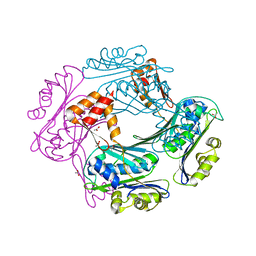 | | Crystal structure of the Bacillus subtilis HSP33 | | Descriptor: | 33 KDA CHAPERONIN, ACETATE ION, ZINC ION | | Authors: | Janda, I.K, Devedjiev, Y, Derewenda, U, Dauter, Z, Bielnicki, J, Cooper, D.R, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-29 | | Release date: | 2004-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of the reduced, Zn2+-bound form of the B. subtilis Hsp33 chaperone and its implications for the activation mechanism.
Structure, 12, 2004
|
|
1ESE
 
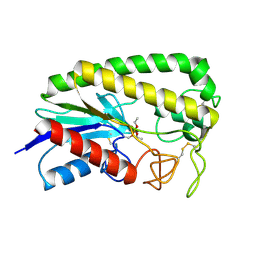 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | DIETHYL PHOSPHONATE, ESTERASE | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
1ESD
 
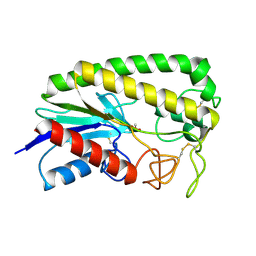 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | ESTERASE, METHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
2JHY
 
 | |
2JI0
 
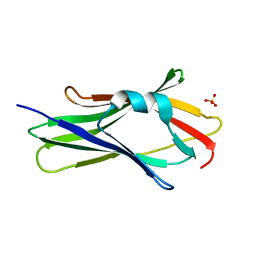 | | CRYSTAL STRUCTURE OF RHOGDI K138Y, K141Y MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Cooper, D.R, Boczek, T, Derewenda, Z.S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein Crystallization by Surface Entropy Reduction: Optimization of the Ser Strategy
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JHT
 
 | | CRYSTAL STRUCTURE OF RHOGDI K135T,K138T,K141T MUTANT | | Descriptor: | LITHIUM ION, RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Cooper, D.R, Grelewska, K, Derewenda, Z.S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Protein Crystallization by Surface Entropy Reduction: Optimization of the Ser Strategy
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JHW
 
 | | CRYSTAL STRUCTURE OF RHOGDI E155A, E157A MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Cooper, D.R, Grelewska, K, Derewenda, Z.S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Crystallization by Surface Entropy Reduction: Optimization of the Ser Strategy
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JHX
 
 | |
2JHZ
 
 | |
2JHS
 
 | | CRYSTAL STRUCTURE OF RHOGDI K135H,K138H,K141H MUTANT | | Descriptor: | CITRATE ANION, RHO GDP-DISSOCIATION INHIBITOR 1 | | Authors: | Cooper, D.R, Zawadzki, M, Derewenda, Z.S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Protein Crystallization by Surface Entropy Reduction: Optimization of the Ser Strategy
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JHV
 
 | |
2JHU
 
 | | CRYSTAL STRUCTURE OF RHOGDI E154A,E155A MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Cooper, D.R, Pinkowska, M, Derewenda, Z.S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protein Crystallization by Surface Entropy Reduction: Optimization of the Ser Strategy
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1KMQ
 
 | | Crystal Structure of a Constitutively Activated RhoA Mutant (Q63L) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Longenecker, K, Read, P, Lin, S.-K, Somlyo, A.P, Nakamoto, R.K, Derewenda, Z.S. | | Deposit date: | 2001-12-17 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a constitutively activated RhoA mutant (Q63L) at 1.55 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KMT
 
 | | Crystal structure of RhoGDI Glu(154,155)Ala mutant | | Descriptor: | Rho GDP-dissociation inhibitor 1 | | Authors: | Mateja, A, Devedjiev, Y, Krowarsh, D, Longenecker, K, Dauter, Z, Otlewski, J, Derewenda, Z.S. | | Deposit date: | 2001-12-17 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The impact of Glu-->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1TGL
 
 | | A SERINE PROTEASE TRIAD FORMS THE CATALYTIC CENTRE OF A TRIACYLGLYCEROL LIPASE | | Descriptor: | TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z.S, Dodson, E.J, Dodson, G.G, Tolley, S.P, Turkenburg, J.P, Christiansen, L, Huge-Jensen, B, Norskov, L, Thim, L. | | Deposit date: | 1990-02-05 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A serine protease triad forms the catalytic centre of a triacylglycerol lipase.
Nature, 343, 1990
|
|
