1Q8R
 
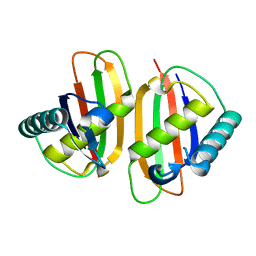 | | Structure of E.coli RusA Holliday junction resolvase | | Descriptor: | Crossover junction endodeoxyribonuclease rusA | | Authors: | Rafferty, J.B, Bolt, E.L, Muranova, T.A, Sedelnikova, S.E, Leonard, P, Pasquo, A, Baker, P.J, Rice, D.W, Sharples, G.J, Lloyd, R.G. | | Deposit date: | 2003-08-22 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | The structure of Escherichia coli RusA endonuclease reveals a new Holliday junction DNA binding fold
Structure, 11, 2003
|
|
1HRD
 
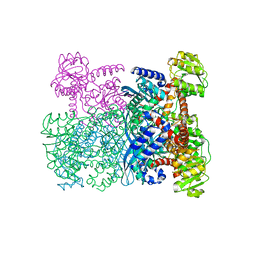 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Britton, K.L, Baker, P.J, Stillman, T.J, Rice, D.W. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures.
Structure, 3, 1995
|
|
6ZZ5
 
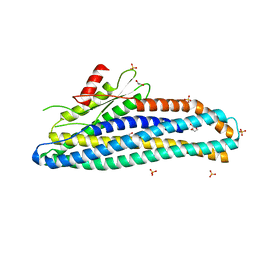 | | Structure of soluble SmhB of the tripartite alpha-pore forming toxin, Smh, from Serratia marcescens. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, SmhB | | Authors: | Churchill-Angus, A.M, Baker, P.J. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Characterisation of a tripartite alpha-pore forming toxin from Serratia marcescens
Sci Rep, 11, 2021
|
|
6ZZH
 
 | |
7A27
 
 | |
7A0G
 
 | |
7A26
 
 | |
1JPU
 
 | | Crystal Structure of Bacillus Stearothermophilus Glycerol Dehydrogenase | | Descriptor: | ZINC ION, glycerol dehydrogenase | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
1JQW
 
 | | THE 2.3 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/HOMOCYSTEINE COMPLEX | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-09 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
1JVI
 
 | | THE 2.2 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/RIBOSILHOMOCYSTEINE COMPLEX | | Descriptor: | (2S)-2-amino-4-[[(2S,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methylsulfanyl]butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ... | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
1ZUW
 
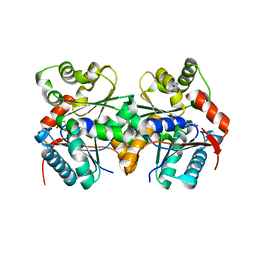 | | Crystal structure of B.subtilis glutamate racemase (RacE) with D-Glu | | Descriptor: | D-GLUTAMIC ACID, glutamate racemase 1 | | Authors: | Ruzheinikov, S.N, Taal, M.A, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2005-06-01 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-Induced Conformational Changes in Bacillus subtilis Glutamate Racemase and Their Implications for Drug Discovery
Structure, 13, 2005
|
|
1JQA
 
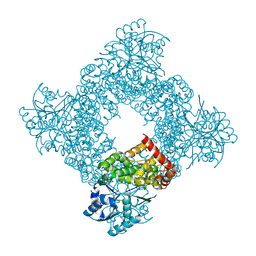 | | Bacillus stearothermophilus glycerol dehydrogenase complex with glycerol | | Descriptor: | GLYCEROL, Glycerol Dehydrogenase, ZINC ION | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-04 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
1L0I
 
 | | Crystal structure of butyryl-ACP I62M mutant | | Descriptor: | Acyl carrier protein, CACODYLATE ION, SODIUM ION, ... | | Authors: | Roujeinikova, A, Baldock, C, Simon, W.J, Gilroy, J, Baker, P.J, Stuitje, A.R, Rice, D.W, Slabas, A.R, Rafferty, J.B. | | Deposit date: | 2002-02-11 | | Release date: | 2003-02-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray Crystallographic Studies on Butyryl-ACP Reveal Flexibility of the Structure around a Putative Acyl Chain Binding Site
Structure, 10, 2002
|
|
1L0H
 
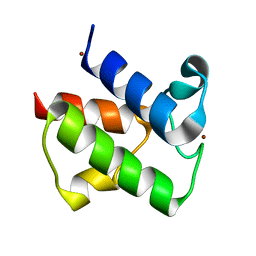 | | CRYSTAL STRUCTURE OF BUTYRYL-ACP FROM E.COLI | | Descriptor: | ACYL CARRIER PROTEIN, ZINC ION | | Authors: | Roujeinikova, A, Baldock, C, Simon, W.J, Gilroy, J, Baker, P.J, Stuitje, A.R, Rice, D.W, Slabas, A.R, Rafferty, J.B. | | Deposit date: | 2002-02-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site
Structure, 10, 2002
|
|
1JQ5
 
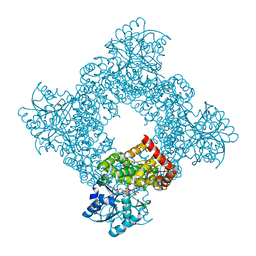 | | Bacillus Stearothermophilus Glycerol dehydrogenase complex with NAD+ | | Descriptor: | Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
2B5W
 
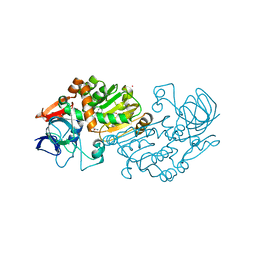 | | Crystal structure of D38C glucose dehydrogenase mutant from Haloferax mediterranei | | Descriptor: | CITRATE ANION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Britton, K.L, Baker, P.J, Fisher, M, Ruzheinikov, S, Gilmour, D.J, Bonete, M.-J, Ferrer, J, Pire, C, Esclapez, J, Rice, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2B5V
 
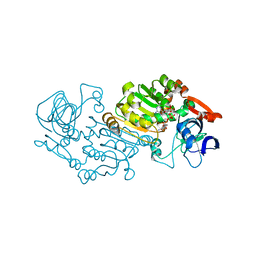 | | Crystal structure of glucose dehydrogenase from Haloferax mediterranei | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, glucose dehydrogenase | | Authors: | Britton, K.L, Baker, P.J, Fisher, M, Ruzheinikov, S, Gilmour, D.J, Bonete, M.-J, Ferrer, J, Pire, C, Esclapez, J, Rice, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2VXY
 
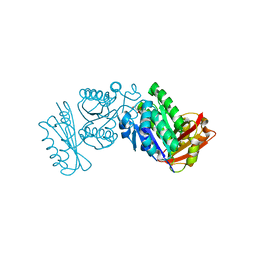 | | The structure of FTsZ from Bacillus subtilis at 1.7A resolution | | Descriptor: | CELL DIVISION PROTEIN FTSZ, CITRIC ACID, POTASSIUM ION | | Authors: | Barynin, V.V, Baker, P.J, Rice, D.W, Sedelnikova, S.E, Haydon, D.J, Stokes, N.R, Ure, R, Galbraith, G, Bennett, J.M, Brown, D.R, Heal, J.R, Sheridan, J.M, Aiwale, S.T, Chauhan, P.K, Srivastava, A, Taneja, A, Collins, I, Errington, J, Czaplewski, L.G. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Inhibitor of Ftsz with Potent and Selective Anti-Staphylococcal Activity.
Science, 321, 2008
|
|
1KTG
 
 | | Crystal Structure of a C. elegans Ap4A Hydrolase Binary Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Diadenosine Tetraphosphate Hydrolase, HYDROXIDE ION, ... | | Authors: | Bailey, S, Sedelnikova, S.E, Blackburn, G.M, Abdelghany, H.M, Baker, P.J, McLennan, A.G, Rafferty, J.B. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of diadenosine tetraphosphate hydrolase from Caenorhabditis elegans in free and binary complex forms
Structure, 10, 2002
|
|
1KKR
 
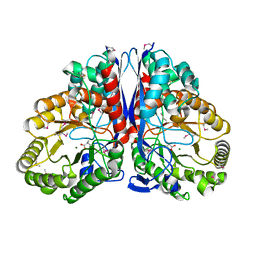 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE CONTAINING (2S,3S)-3-METHYLASPARTIC ACID | | Descriptor: | (2S,3S)-3-methyl-aspartic acid, 3-METHYLASPARTATE AMMONIA-LYASE, MAGNESIUM ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, K, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
3FYM
 
 | | The 1A structure of YmfM, a putative DNA-binding membrane protein from Staphylococcus aureus | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Xu, L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2009-01-22 | | Release date: | 2010-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The 1A structure of YmfM, a putative DNA-binding membrane protein from Staphylococcus aureus
To be Published
|
|
1KKO
 
 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE | | Descriptor: | 3-METHYLASPARTATE AMMONIA-LYASE, SULFATE ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, Y, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
1KT9
 
 | | Crystal Structure of C. elegans Ap4A Hydrolase | | Descriptor: | Diadenosine Tetraphosphate Hydrolase | | Authors: | Bailey, S, Sedelnikova, S.E, Blackburn, G.M, Abdelghany, H.M, Baker, P.J, McLennan, A.G, Rafferty, J.B. | | Deposit date: | 2002-01-15 | | Release date: | 2002-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of diadenosine tetraphosphate hydrolase from Caenorhabditis elegans in free and binary complex forms
Structure, 10, 2002
|
|
1IGW
 
 | | Crystal Structure of the Isocitrate Lyase from the A219C mutant of Escherichia coli | | Descriptor: | Isocitrate lyase, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Britton, K.L, Abeysinghe, I.S.B, Baker, P.J, Barynin, V, Diehl, P, Langridge, S.J, McFadden, B.A, Sedelnikova, S.E, Stillman, T.J, Weeradechapon, K, Rice, D.W. | | Deposit date: | 2001-04-18 | | Release date: | 2001-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure and domain organization of Escherichia coli isocitrate lyase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1J98
 
 | | The 1.2 Angstrom Structure of Bacillus subtilis LuxS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-05-24 | | Release date: | 2001-06-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A Structure of a Novel Quorum-Sensing Protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
