1L4K
 
 | | Crystal Structure of CobT complexed with 3,4-dimethylaniline and nicotinate mononucleotide | | Descriptor: | 3,4-DIMETHYLANILINE, NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica.
J.Biol.Chem., 277, 2002
|
|
1JHR
 
 | | Three-dimensional Structure of CobT in Complex with Reaction Products of 2-hydroxypurine and NaMN | | Descriptor: | N7-(5'-PHOSPHO-ALPHA-RIBOSYL)-2-HYDROXYPURINE, NICOTINIC ACID, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1L4L
 
 | | Crystal Structure of CobT complexed with 2,5-dimethylaniline and nicotinate mononucleotide | | Descriptor: | 2,5-DIMETHYLANILINE, NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica.
J.Biol.Chem., 277, 2002
|
|
1L4M
 
 | | Crystal Structure of CobT complexed with 2-amino-p-cresol and nicotinate mononucleotide | | Descriptor: | 2-AMINO-P-CRESOL, NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica.
J.Biol.Chem., 277, 2002
|
|
1JHA
 
 | | Structural Investigation of the Biosynthesis of Alternative Lower Ligands for Cobamides by Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase (CobT) from Salmonella enterica | | Descriptor: | ALPHA-ADENOSINE MONOPHOSPHATE, NICOTINIC ACID, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1JHM
 
 | | Three-dimensional Structure of CobT in Complex with 5-methylbenzimidazole | | Descriptor: | 5-METHYLBENZIMIDAZOLE, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1JHU
 
 | | Three-dimensional Structure of CobT in Complex with p-cresol | | Descriptor: | Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase, P-CRESOL, PHOSPHATE ION | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1L4H
 
 | | Crystal Structure of CobT complexed with indole and nicotinate mononucleotide | | Descriptor: | INDOLE, NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica.
J.Biol.Chem., 277, 2002
|
|
3HNE
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to the effectors TTP and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7L02
 
 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to one copy of domain-swapped antibody 2G12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L09
 
 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound domain-swapped antibody 2G12 from masked 3D refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L6M
 
 | |
7L6O
 
 | | Cryo-EM structure of HIV-1 Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 - gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2020-12-23 | | Release date: | 2021-04-14 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
3HNC
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to the effector TTP | | Descriptor: | MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, SULFATE ION, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7LUA
 
 | | Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848 SOSIP gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2021-02-21 | | Release date: | 2021-03-17 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7LU9
 
 | | Cryo-EM structure of DH851.3 bound to HIV-1 CH505 Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2021-02-21 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L06
 
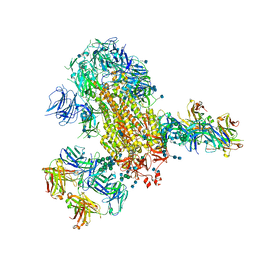 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to two copies of domain-swapped antibody 2G12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
3HNF
 
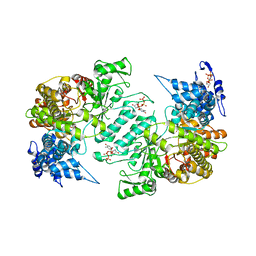 | | Crystal structure of human ribonucleotide reductase 1 bound to the effectors TTP and dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1PP2
 
 | |
2XT0
 
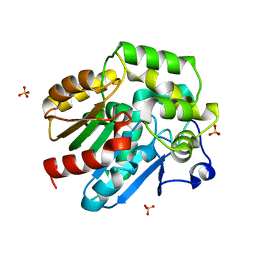 | | Dehalogenase DPpA from Plesiocystis pacifica SIR-I | | Descriptor: | HALOALKANE DEHALOGENASE, SULFATE ION | | Authors: | Bogdanovic, X, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-10-02 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cloning, Functional Expression, Biochemical Characterization, and Structural Analysis of a Haloalkane Dehalogenase from Plesiocystis Pacifica Sir-1.
Appl.Microbiol.Biotechnol., 91, 2011
|
|
1BCD
 
 | |
3GEC
 
 | |
3GDI
 
 | |
3HND
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to the effector TTP and substrate GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2F9Y
 
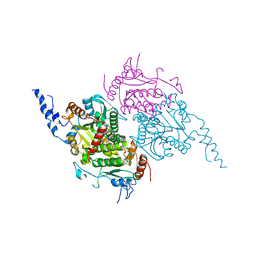 | | The Crystal Structure of The Carboxyltransferase Subunit of ACC from Escherichia coli | | Descriptor: | Acetyl-CoA carboxylase, Carboxyltransferase alpha chain, Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta, ... | | Authors: | Bilder, P.W. | | Deposit date: | 2005-12-06 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure of the Carboxyltransferase Component of Acetyl-CoA Carboxylase Reveals a Zinc-Binding Motif Unique to the Bacterial Enzyme(,).
Biochemistry, 45, 2006
|
|
