1NPR
 
 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN C222(1) | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzhan, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
2PSX
 
 | |
1LU0
 
 | | Atomic Resolution Structure of Squash Trypsin Inhibitor: Unexpected Metal Coordination | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Thaimattam, R, Tykarska, E, Bierzynski, A, Sheldrick, G.M, Jaskolski, M. | | Deposit date: | 2002-05-21 | | Release date: | 2002-08-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution structure of squash trypsin inhibitor: unexpected metal coordination.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (UBIQUINONE-2 COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB.
Structure, 5, 1997
|
|
2PGT
 
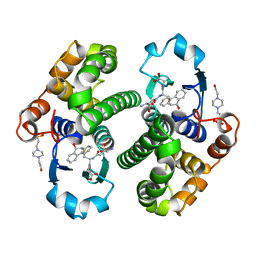 | | CRYSTAL STRUCTURE OF HUMAN GLUTATHIONE S-TRANSFERASE P1-1[V104] COMPLEXED WITH (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, ... | | Authors: | Ji, X. | | Deposit date: | 1997-02-17 | | Release date: | 1997-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate-binding site and location of a potential non-substrate-binding site in a class pi glutathione S-transferase.
Biochemistry, 36, 1997
|
|
1HT1
 
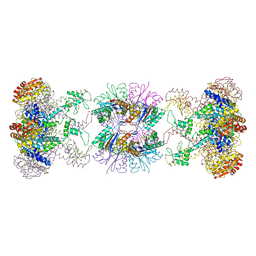 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1HT2
 
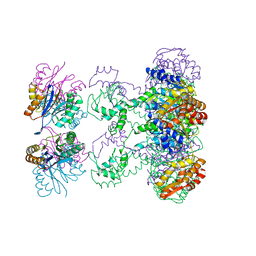 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1AIJ
 
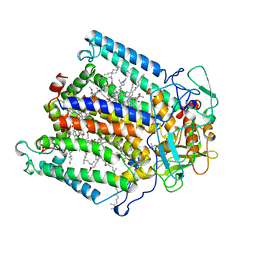 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE CHARGE-NEUTRAL DQAQB STATE | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Stowell, M.H.B, Mcphillips, T.M, Soltis, S.M, Rees, D.C, Abresch, E, Feher, G. | | Deposit date: | 1997-04-18 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer.
Science, 276, 1997
|
|
1BIT
 
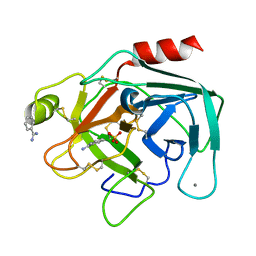 | |
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1AMH
 
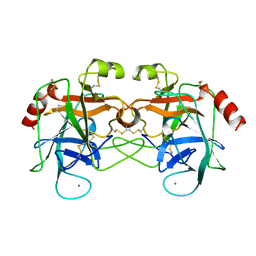 | | UNCOMPLEXED RAT TRYPSIN MUTANT WITH ASP 189 REPLACED WITH SER (D189S) | | Descriptor: | ANIONIC TRYPSIN, CALCIUM ION | | Authors: | Szabo, E, Bocskei, Z.S, Naray-Szabo, G, Graf, L. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three-dimensional structure of Asp189Ser trypsin provides evidence for an inherent structural plasticity of the protease.
Eur.J.Biochem., 263, 1999
|
|
1AIG
 
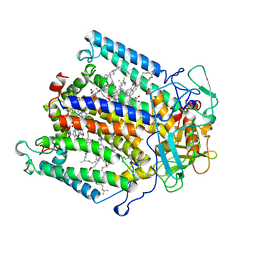 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE D+QB-CHARGE SEPARATED STATE | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Stowell, M.H.B, Mcphillips, T.M, Soltis, S.M, Rees, D.C, Abresch, E, Feher, G. | | Deposit date: | 1997-04-17 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer.
Science, 276, 1997
|
|
4AIG
 
 | | ADAMALYSIN II WITH PHOSPHONATE INHIBITOR | | Descriptor: | ADAMALYSIN II, CALCIUM ION, N-[(FURAN-2-YL)CARBONYL]-(S)-LEUCYL-(R)-[1-AMINO-2(1H-INDOL-3-YL)ETHYL]-PHOSPHONIC ACID, ... | | Authors: | Pochetti, G, Mazza, F, Gavuzzo, E, Cirilli, M. | | Deposit date: | 1997-10-16 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2 angstrom X-ray structure of adamalysin II complexed with a peptide phosphonate inhibitor adopting a retro-binding mode.
FEBS Lett., 418, 1997
|
|
1AZN
 
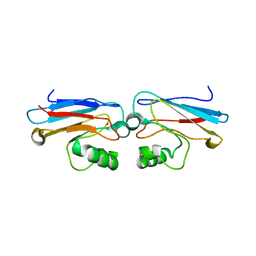 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT PHE114ALA FROM PSEUDOMONAS AERUGINOSA AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Pascher, T, Nar, H. | | Deposit date: | 1994-05-27 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the azurin mutant Phe114Ala from Pseudomonas aeruginosa at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1CTI
 
 | |
3ZI5
 
 | | Crystal STRUCTURE OF RESTRICTION ENDONUCLEASE BFII C-TERMINAL RECOGNITION DOMAIN IN COMPLEX WITH COGNATE DNA | | Descriptor: | 5'-D(*AP*GP*CP*AP*CP*TP*GP*GP*GP*TP*CP*GP)-3', 5'-D(*CP*GP*AP*CP*CP*CP*AP*GP*TP*GP*CP*TP)-3', RESTRICTION ENDONUCLEASE | | Authors: | Golovenko, D, Manakova, E, Zakrys, L, Zaremba, M, Sasnauskas, G, Grazulis, S, Siksnys, V. | | Deposit date: | 2013-01-03 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insight Into the Specificity of the B3 DNA-Binding Domains Provided by the Co-Crystal Structure of the C-Terminal Fragment of Bfii Restriction Enzyme
Nucleic Acids Res., 42, 2014
|
|
1PPC
 
 | | GEOMETRY OF BINDING OF THE BENZAMIDINE-AND ARGININE-BASED INHIBITORS N-ALPHA-(2-NAPHTHYL-SULPHONYL-GLYCYL)-DL-P-AMIDINOPHENYLALANYL-PIPERIDINE (NAPAP) AND (2R,4R)-4-METHYL-1-[N-ALPHA-(3-METHYL-1,2,3,4-TETRAHYDRO-8-QUINOLINESULPHONYL)-L-ARGINYL]-2-PIPERIDINE CARBOXYLIC ACID (MQPA) TO HUMAN ALPHA-THROMBIN: X-RAY CRYSTALLOGRAPHIC DETERMINATION OF THE NAPAP-TRYPSIN COMPLEX AND MODELING OF NAPAP-THROMBIN AND MQPA-THROMBIN | | Descriptor: | 1-[N-(naphthalen-2-ylsulfonyl)glycyl-4-carbamimidoyl-D-phenylalanyl]piperidine, CALCIUM ION, TRYPSIN | | Authors: | Bode, W, Turk, D. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Geometry of binding of the benzamidine- and arginine-based inhibitors N alpha-(2-naphthyl-sulphonyl-glycyl)-DL-p-amidinophenylalanyl-pipe ridine (NAPAP) and (2R,4R)-4-methyl-1-[N alpha-(3-methyl-1,2,3,4-tetrahydr quinolinesulphonyl)-L-arginyl]-2-piperidine carboxylic acid (MQPA) to human alpha-thrombin.X-ray crystallographic determination of the NAPAP-trypsin complex and modeling of NAPAP-thrombin and MQPA-thrombin.
Eur.J.Biochem., 193, 1990
|
|
1LKT
 
 | | CRYSTAL STRUCTURE OF THE HEAD-BINDING DOMAIN OF PHAGE P22 TAILSPIKE PROTEIN | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage.
J.Mol.Biol., 267, 1997
|
|
1SIJ
 
 | | Crystal structure of the Aldehyde Dehydrogenase (a.k.a. AOR or MOP) of Desulfovibrio gigas covalently bound to [AsO3]- | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ARSENITE, Aldehyde oxidoreductase, ... | | Authors: | Boer, D.R, Thapper, A, Brondino, C.D, Romao, M.J, Moura, J.J.G. | | Deposit date: | 2004-03-01 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure and EPR Spectra of "Arsenite-Inhibited" Desulfovibriogigas Aldehyde Dehydrogenase: A Member of the Xanthine Oxidase Family
J.Am.Chem.Soc., 126, 2004
|
|
1MMB
 
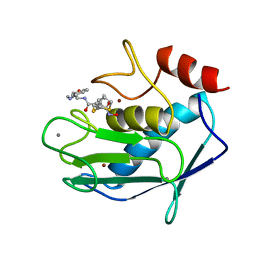 | | COMPLEX OF BB94 WITH THE CATALYTIC DOMAIN OF MATRIX METALLOPROTEINASE-8 | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, MATRIX METALLOPROTEINASE-8, ... | | Authors: | Bode, W, Grams, F. | | Deposit date: | 1995-08-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination and analysis of human neutrophil collagenase complexed with a hydroxamate inhibitor.
Biochemistry, 34, 1995
|
|
1PSS
 
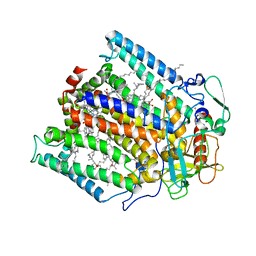 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
2GSS
 
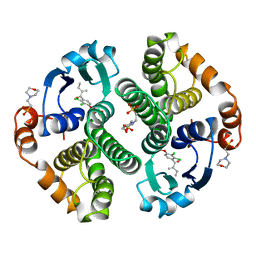 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 IN COMPLEX WITH ETHACRYNIC ACID | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ETHACRYNIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A.J, Rossjohn, J, Parker, M.W. | | Deposit date: | 1996-10-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of the human Pi class glutathione transferase P1-1 in complex with the inhibitor ethacrynic acid and its glutathione conjugate.
Biochemistry, 36, 1997
|
|
1PST
 
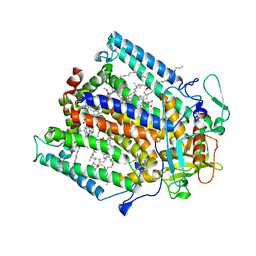 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
2GLR
 
 | |
1PGT
 
 | |
