7JHL
 
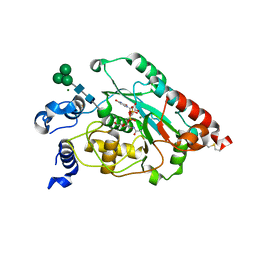 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with UDP-N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
8XMS
 
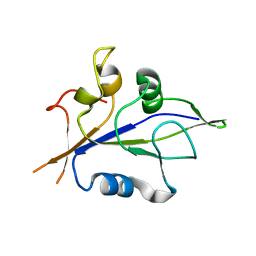 | |
3K51
 
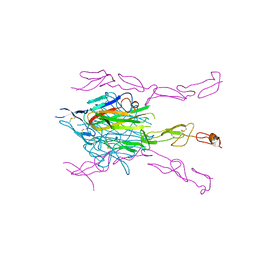 | | Crystal Structure of DcR3-TL1A complex | | Descriptor: | Decoy receptor 3, Tumor necrosis factor ligand superfamily member 15, secreted form | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2009-10-06 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
6LJJ
 
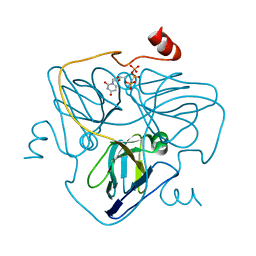 | | Swine dUTPase in complex with alpha,beta-iminodUTP and magnesium ion | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial isoform 1, ... | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6LAU
 
 | | the wildtype SAM-VI riboswitch bound to SAH | | Descriptor: | CESIUM ION, GUANOSINE-5'-TRIPHOSPHATE, RNA (54-MER), ... | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
6LBS
 
 | | Crystal structure of yeast Stn1 | | Descriptor: | KLLA0C11825p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBR
 
 | | Crystal structure of yeast Cdc13 and ssDNA | | Descriptor: | KLLA0F20922p, Telomere single-strand DNA | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBT
 
 | | Crystal structure of yeast Cdc13 and Stn1 | | Descriptor: | GLYCEROL, KLLA0C11825p, KLLA0F20922p,KLLA0F20922p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBU
 
 | | Crystal structure of yeast Stn1 and Ten1 | | Descriptor: | KLLA0C11825p, KLLA0E09417p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LAX
 
 | | the mutant SAM-VI riboswitch (U6C) bound to SAM | | Descriptor: | RNA (55-MER), S-ADENOSYLMETHIONINE, U1 small nuclear ribonucleoprotein A | | Authors: | Sun, A, Ren, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
6LAZ
 
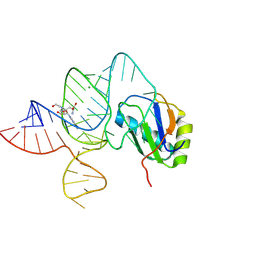 | | the wildtype SAM-VI riboswitch bound to a N-mustard SAM analog M1 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(2-hydroxyethyl)amino]-2-azaniumyl-butanoate, MAGNESIUM ION, RNA (55-MER), ... | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
6LAS
 
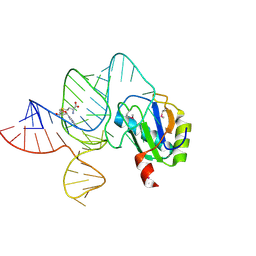 | | the wildtype SAM-VI riboswitch bound to SAM | | Descriptor: | RNA (55-MER), S-ADENOSYLMETHIONINE, U1 small nuclear ribonucleoprotein A | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
7F4N
 
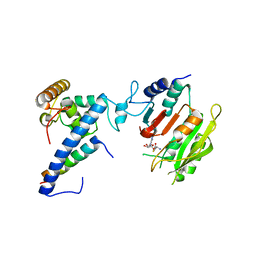 | | Crystal structure of SAH-bound MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYL-L-HOMOCYSTEINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4R
 
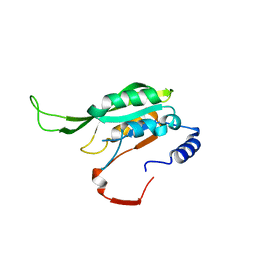 | | Crystal structure of MTA1 | | Descriptor: | MT-a70 family protein | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4P
 
 | | Crystal structure of SAM-bound MTA1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYLMETHIONINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4S
 
 | | Crystal structure of TthMTA1-PteMTA9 complex | | Descriptor: | MT-a70 family protein, MTA9 | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4O
 
 | | Crystal structure of MTA1-p2 complex | | Descriptor: | MT-a70 family protein, Transmembrane protein, putative | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4Q
 
 | | Crystal structure of SAH-bound MTA1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYL-L-HOMOCYSTEINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4L
 
 | | Crystal structure of MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, Transmembrane protein, putative, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4M
 
 | | Crystal structure of SAM-bound MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYLMETHIONINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
6LIS
 
 | | ASFV dUTPase in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
7F4T
 
 | |
6LJ3
 
 | |
6LJO
 
 | | African swine fever virus dUTPase | | Descriptor: | E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
7DK0
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW05 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW05 heavy chain, MW05 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
