6A96
 
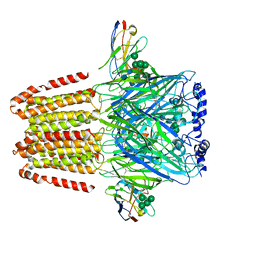 | | Cryo-EM structure of the human alpha5beta3 GABAA receptor in complex with GABA and Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyric acid receptor subunit alpha-5, ... | | Authors: | Liu, S, Xu, L, Guan, F, Liu, Y.T, Cui, Y, Zhang, Q, Bi, G.Q, Zhou, Z.H, Zhang, X, Ye, S. | | Deposit date: | 2018-07-11 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM structure of the human alpha 5 beta 3 GABAAreceptor.
Cell Res., 28, 2018
|
|
3UUO
 
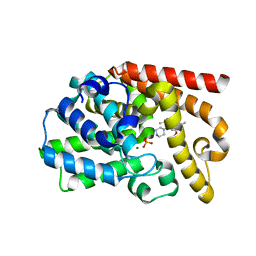 | | The discovery of potent, selectivity, and orally bioavailable pyrozoloquinolines as PDE10 inhibitors for the treatment of Schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(piperazin-1-yl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ho, G.D, Yang, S, Smotryski, J, Bercovici, A, Nechuta, T, Smith, E.M, McElroy, W, Tan, Z, Tulshian, D, Mckittrick, B, Greenlee, W.J, Hruza, A, Xiao, L, Rindgen, D, Guzzi, M, Zhang, X, Bleickardt, C, Mullins, D, Hodgson, R. | | Deposit date: | 2011-11-28 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The discovery of potent, selective, and orally active pyrazoloquinolines as PDE10A inhibitors for the treatment of Schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4HIL
 
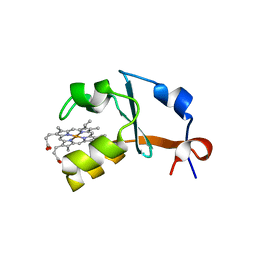 | | 1.25A Resolution Structure of Rat Type B Cytochrome b5 | | Descriptor: | Cytochrome b5 type B, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Parthasarathy, S, Sun, N, Terzyan, S, Zhang, X, Rivera, M, Kuczera, K, Benson, D.R. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25A Resolution Structure of Rat Type B Cytochrome b5
To be Published
|
|
8W9D
 
 | |
8W9C
 
 | |
8I4H
 
 | | Omicron spike variant BA.1 with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8W9F
 
 | |
8W9E
 
 | |
4HIN
 
 | | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L) | | Descriptor: | COPPER (II) ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Parthasarathy, S, Sun, N, Terzyan, S, Zhang, X, Rivera, M, Kuczera, K, Benson, D.R. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L)
To be Published
|
|
5X5S
 
 | |
4IP3
 
 | | Complex structure of OspI and Ubc13 | | Descriptor: | ORF169b, Ubiquitin-conjugating enzyme E2 N | | Authors: | Fu, P, Jin, M, Zhang, X, Xu, L, Xia, Z, Zhu, Y. | | Deposit date: | 2013-01-09 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Analysis of Ubc13 Inactivation
To be Published
|
|
7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | Descriptor: | NB_1B11, Spike protein S1, ZINC ION | | Authors: | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
4JPN
 
 | | Bacteriophage phiX174 H protein residues 143-221 | | Descriptor: | Minor spike protein H | | Authors: | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
4JPP
 
 | | Bacteriophage phiX174 H protein residues 143-282 | | Descriptor: | Minor spike protein H | | Authors: | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
8I35
 
 | | Acyl-ACP synthetase structure bound to oleic acid | | Descriptor: | Acyl-acyl carrier protein synthetase, OLEIC ACID | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Acyl-ACP synthetase structure bound to oleic acid
To Be Published
|
|
8I51
 
 | | Acyl-ACP synthetase structure bound to AMP-MC7 | | Descriptor: | 7-methoxy-7-oxidanylidene-heptanoic acid, ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Acyl-ACP synthetase structure bound to AMP-MC7
To Be Published
|
|
8I3I
 
 | | Acyl-ACP synthetase structure bound to AMP-PNP in the presence of MgCl2 | | Descriptor: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-17 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Acyl-ACP synthetase structure bound to AMP-PNP in the presence of MgCl2
To Be Published
|
|
8I6M
 
 | | Acyl-ACP synthetase structure bound to AMP-C18:1 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, MAGNESIUM ION, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Acyl-ACP synthetase structure bound to AMP-C18:1
To Be Published
|
|
8HZX
 
 | | Acyl-ACP synthetase structure-2 | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Acyl-ACP synthetase structure-2
To Be Published
|
|
8I49
 
 | | Acyl-ACP synthetase structure bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Acyl-ACP synthetase structure bound to ATP.
To Be Published
|
|
8I8D
 
 | | Acyl-ACP synthetase structure bound to MC7-ACP | | Descriptor: | 7-methoxy-7-oxidanylidene-heptanoic acid, ADENOSINE MONOPHOSPHATE, Acyl carrier protein, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Acyl-ACP synthetase structure bound to MC7-ACP
To Be Published
|
|
8I8E
 
 | | Acyl-ACP synthetase structure bound to C18:1-ACP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, Acyl carrier protein, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Acyl-ACP synthetase structure bound to C18:1-ACP
To Be Published
|
|
8H2I
 
 | | Near-atomic structure of five-fold averaged PBCV-1 capsid | | Descriptor: | MCPv1, MCPv2, MCPv3, ... | | Authors: | Shao, Q, Agarkova, I.V, Noel, E.A, Dunigan, D.D, Liu, Y, Wang, A, Guo, M, Xie, L, Zhao, X, Rossmann, M.G, Van Etten, J.L, Klose, T, Fang, Q. | | Deposit date: | 2022-10-06 | | Release date: | 2022-11-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Near-atomic, non-icosahedrally averaged structure of giant virus Paramecium bursaria chlorella virus 1.
Nat Commun, 13, 2022
|
|
8JYL
 
 | | Acyl-ACP Synthetase structure bound to C10-AMS | | Descriptor: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-decanoylsulfamate | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Acyl-ACP Synthetase structure bound to C10-AMS
To Be Published
|
|
8HSY
 
 | | Acyl-ACP Synthetase structure | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Acyl-ACP Synthetase structure
To Be Published
|
|
