6P88
 
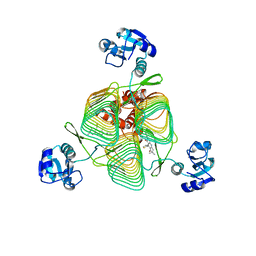 | | E.coli LpxD in complex with compound 6 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, N-{3-[(furan-2-carbonyl)amino]phenyl}-2,3-dihydro-1,4-benzodioxine-6-carboxamide, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P85
 
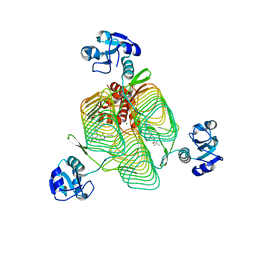 | | E.coli LpxD in complex with compound 3 | | Descriptor: | 1-(2-ethoxyphenyl)-4-[(4-methylphenyl)sulfonyl]-1H-1,2,3-triazol-5-amine, 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P89
 
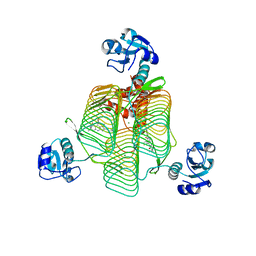 | | E.coli LpxD in complex with compound 7 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, N-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-(8-methyl[1]benzopyrano[4,3-c]pyrazol-1(4H)-yl)acetamide, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P87
 
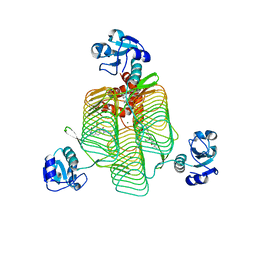 | | E.coli LpxD in complex with compound 5 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, N-[(R)-(1H-indol-3-yl)(thiophen-2-yl)methyl]-2-(2-methoxyphenoxy)-N-methylacetamide, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P8B
 
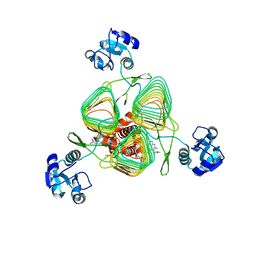 | | E.coli LpxD in complex with peptide FITC-RJPXD33 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, FITC-RJPXD33, MAGNESIUM ION, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P84
 
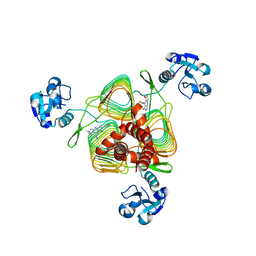 | | E.coli LpxD in complex with compound 2o | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P8A
 
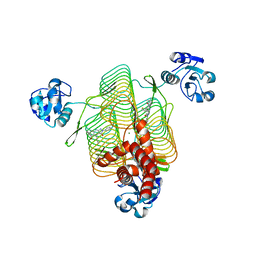 | | E.coli LpxD in complex with compound 8.1 | | Descriptor: | 2-[4,6-dimethyl-3-(1H-pyrrol-1-yl)-1H-pyrazolo[3,4-b]pyridin-1-yl]-N-[3-(morpholin-4-yl)propyl]acetamide, 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
6P83
 
 | | E.coli LpxD in complex with compound 1o | | Descriptor: | 4-(2-chlorophenyl)-3-hydroxy-7,7-dimethyl-2-phenyl-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
8ZNJ
 
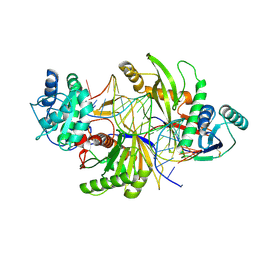 | | Cryo-EM structure of a short prokaryotic Argonaute system from archaeon Suldolobus islandicus | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*CP*AP*GP*GP*GP*TP*AP*TP*CP*TP*AP*AP*GP*CP*TP*TP*TP*GP*AP*A)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Dai, Z.K, Guan, Z.Y, Han, W.Y, Zou, T.T. | | Deposit date: | 2024-05-27 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mechanistic insights into the activation of a short prokaryotic argonaute system from archaeon Sulfolobus islandicus.
Nucleic Acids Res., 53, 2025
|
|
2MK5
 
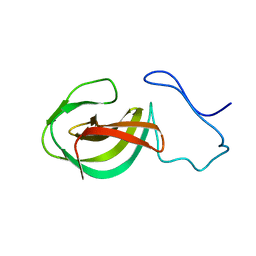 | | Solution structure of a protein domain | | Descriptor: | Endolysin | | Authors: | Feng, Y, Gu, J. | | Deposit date: | 2014-01-24 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
7WK5
 
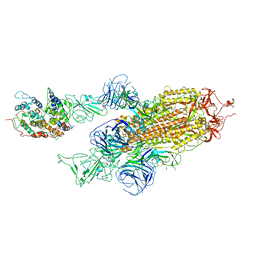 | | Cryo-EM structure of Omicron S-ACE2, C2 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Han, W.Y, Wang, Y.F. | | Deposit date: | 2022-01-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Molecular basis of SARS-CoV-2 Omicron variant receptor engagement and antibody evasion and neutralization
Biorxiv, 2022
|
|
7WK4
 
 | |
7WK6
 
 | |
6L6X
 
 | | The structure of ScoE with substrate | | Descriptor: | (3~{R})-3-(2-hydroxy-2-oxoethylamino)butanoic acid, D(-)-TARTARIC ACID, FE (II) ION, ... | | Authors: | Chen, T.Y, Chen, J, Zhou, J, Chang, W. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Pathway from N-Alkylglycine to Alkylisonitrile Catalyzed by Iron(II) and 2-Oxoglutarate-Dependent Oxygenases.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6L86
 
 | | The structure of SfaA | | Descriptor: | (2S)-2-hydroxybutanedioic acid, D-MALATE, FE (II) ION, ... | | Authors: | Chen, T.Y, Chen, J, Zhou, J, Chang, W. | | Deposit date: | 2019-11-05 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Pathway from N-Alkylglycine to Alkylisonitrile Catalyzed by Iron(II) and 2-Oxoglutarate-Dependent Oxygenases.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7O70
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4R,7S)-12-chloro-14-fluoro-13-(2-fluoro-6-hydroxyphenyl)-4-methyl-10-oxa-2,5,16,18-tetrazatetracyclo[9.7.1.0^(2,7).0^(15,19)]nonadeca-1(18),11,13,15(19),16-pentaen-5-en-1-one-yl]prop-2, CALCIUM ION, GTPase KRas, ... | | Authors: | Phillips, C. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
7OO7
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(6aS)-3-chloro-2-(5-methyl-1H-indazol-4-yl)-5,6,6a,7,9,10-hexahydro-8H-pyrazino[1',2':5,6][1,5]oxazocino[4,3,2-de]quinazolin-8-yl]-2-propen-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Phillips, C. | | Deposit date: | 2021-05-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
8ZB6
 
 | |
8ZH6
 
 | |
7WK2
 
 | | SARS-CoV-2 Omicron S-close | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WKA
 
 | |
7WK9
 
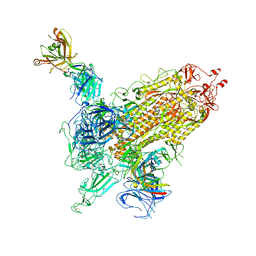 | |
7WK8
 
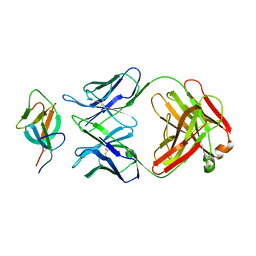 | |
7WVO
 
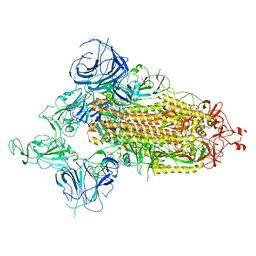 | | SARS-CoV-2 Omicron S-open-2 | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WVN
 
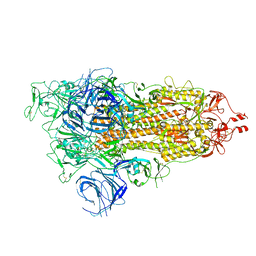 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
