6LDF
 
 | |
6LDE
 
 | |
6LDG
 
 | |
6LIU
 
 | | Crystal structure of apo Tyrosine decarboxylase | | Descriptor: | Tyrosine/DOPA decarboxylase 2 | | Authors: | Yu, J, Wang, H, Yao, M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures clarify cofactor binding of plant tyrosine decarboxylase.
Biochem.Biophys.Res.Commun., 2019
|
|
4EGX
 
 | | Crystal structure of KIF1A CC1-FHA tandem | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Kinesin-like protein KIF1A | | Authors: | Yu, J, Huo, L, Yue, Y, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-02 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
9KRH
 
 | | human creatine transporter | | Descriptor: | CHLORIDE ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SODIUM ION, ... | | Authors: | Yu, J, Ge, J.P, Chen, J.H. | | Deposit date: | 2024-11-27 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transport and inhibition mechanisms of human creatine transporter.
Cell Discov, 11, 2025
|
|
9KRI
 
 | | human creatine transporter | | Descriptor: | 3-carbamimidamidopropanoic acid, CHLORIDE ION, SODIUM ION, ... | | Authors: | Yu, J, Ge, J.P, Chen, J.H. | | Deposit date: | 2024-11-27 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Transport and inhibition mechanisms of human creatine transporter.
Cell Discov, 11, 2025
|
|
9KR7
 
 | | human creatine transporter | | Descriptor: | CHLORIDE ION, Sodium- and chloride-dependent creatine transporter 1 | | Authors: | Yu, J, Ge, J.P, Chen, J.H. | | Deposit date: | 2024-11-27 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Transport and inhibition mechanisms of human creatine transporter.
Cell Discov, 11, 2025
|
|
4YMX
 
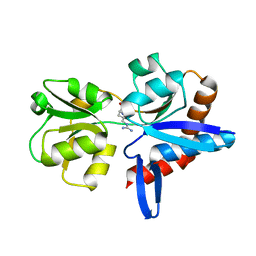 | |
4YMV
 
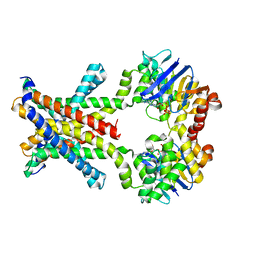 | | Crystal structure of an amino acid ABC transporter with ATPs | | Descriptor: | ABC-type amino acid transport system, permease component, ABC-type polar amino acid transport system, ... | | Authors: | Ge, J, Yu, J, Yang, M. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis for substrate specificity of an amino acid ABC transporter
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YMU
 
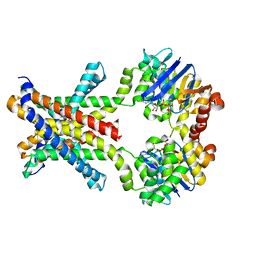 | | Crystal structure of an amino acid ABC transporter complex with arginines and ATPs | | Descriptor: | ABC-type amino acid transport system, permease component, ABC-type polar amino acid transport system, ... | | Authors: | Ge, J, Yu, J, Yang, M. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural basis for substrate specificity of an amino acid ABC transporter
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YMS
 
 | | Crystal structure of an amino acid ABC transporter | | Descriptor: | ABC-type amino acid transport system, permease component, ABC-type polar amino acid transport system, ... | | Authors: | Ge, J, Yu, J, Yang, M. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for substrate specificity of an amino acid ABC transporter
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YMW
 
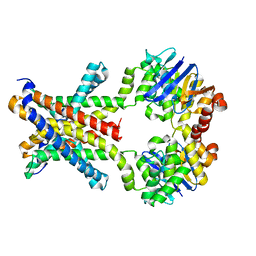 | | Crystal structure of an amino acid ABC transporter with histidines | | Descriptor: | ABC-type amino acid transport system, permease component, ABC-type polar amino acid transport system, ... | | Authors: | Ge, J, Yu, J, Yang, M. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural basis for substrate specificity of an amino acid ABC transporter
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YMT
 
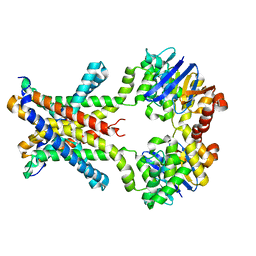 | | Crystal structure of an amino acid ABC transporter complex with arginines | | Descriptor: | ABC-type amino acid transport system, permease component, ABC-type polar amino acid transport system, ... | | Authors: | Ge, J, Yu, J, Yang, M. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Structural basis for substrate specificity of an amino acid ABC transporter
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8W4A
 
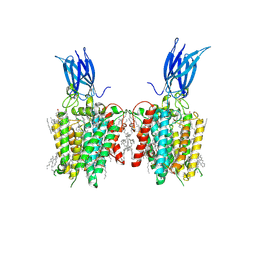 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-08-23 | | Release date: | 2024-10-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure and mechanism of lysosome transmembrane acetylation by HGSNAT.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9IQG
 
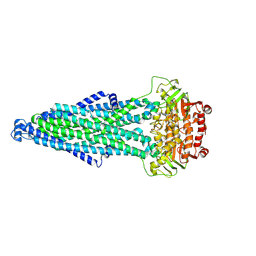 | | Cryo-EM structure of MsRv1273c/72c from Mycobacterium smegmatis in the ATP|ADP+Vi-bound Occ (Vi) state | | Descriptor: | ABC transporter transmembrane region, ABC transporter, ATP-binding protein, ... | | Authors: | Lan, Y, Yu, J, Li, J. | | Deposit date: | 2024-07-12 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of a mycobacterial isoniazid efflux pump MsRv1273c/72c with a degenerate nucleotide-binding site.
Nat Commun, 16, 2025
|
|
9KWI
 
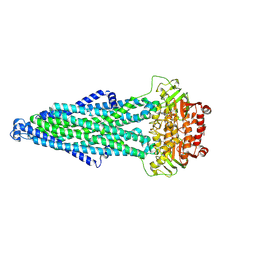 | | Cryo-EM structure of MsRv1273c/72c(E553Q) mutant from Mycobacterium smegmatis in the ATP-bound Occ state | | Descriptor: | ABC transporter, ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lan, Y, Yu, J, Li, J. | | Deposit date: | 2024-12-05 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure and mechanism of a mycobacterial isoniazid efflux pump MsRv1273c/72c with a degenerate nucleotide-binding site.
Nat Commun, 16, 2025
|
|
3CTF
 
 | | Crystal structure of oxidized GRX2 | | Descriptor: | Glutaredoxin-2 | | Authors: | Yu, J, Teng, Y.B, Zhou, C.Z. | | Deposit date: | 2008-04-14 | | Release date: | 2008-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the different activities of yeast Grx1 and Grx2.
Biochim.Biophys.Acta, 1804, 2010
|
|
3HPM
 
 | |
8V8K
 
 | | Crystal Structure of Nanobody NbE | | Descriptor: | Nanobody NbE | | Authors: | Koehl, A, Manglik, A, Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Steyaert, J, Ballet, S, Boland, A, Stoeber, M. | | Deposit date: | 2023-12-05 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of mu-opioid receptor targeting by a nanobody antagonist.
Nat Commun, 15, 2024
|
|
3HPK
 
 | |
3TFM
 
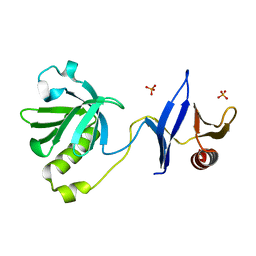 | | Myosin X PH1N-PH2-PH1C tandem | | Descriptor: | Myosin X, PHOSPHATE ION | | Authors: | Yu, J, Lu, Q, Yan, J, Wei, Z, Zhang, M. | | Deposit date: | 2011-08-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis of the myosin X PH1N-PH2-PH1C tandem as a specific and acute cellular PI(3,4,5)P3 sensor
MOLECULAR BIOLOGY OF THE CELL, 22, 2011
|
|
3KYQ
 
 | |
5Y56
 
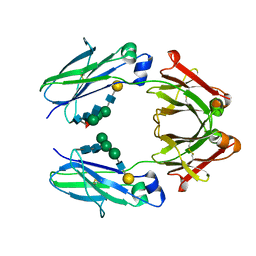 | | Fc mutant (K392D/K409D/D399K) | | Descriptor: | Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[beta-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ye, S, Xu, T, Yu, J, Wang, X, Xu, T, Jin, Q, Duan, J, Wu, J, Wu, H. | | Deposit date: | 2017-08-07 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | A rational approach to enhancing antibody Fc homodimer formation for robust production of antibody mixture in a single cell line
J. Biol. Chem., 292, 2017
|
|
6M16
 
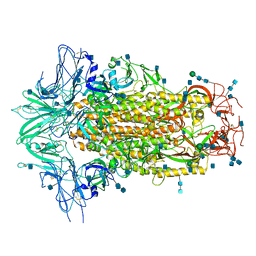 | | Cryo-EM structures of SADS-CoV spike glycoproteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Yu, J, Qiao, S, Guo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
