5A4H
 
 | | Solution structure of the lipid droplet anchoring peptide of CGI-58 bound to DPC micelles | | Descriptor: | 1-ACYLGLYCEROL-3-PHOSPHATE O-ACYLTRANSFERASE ABHD5 | | Authors: | Boeszoermenyi, A, Arthanari, H, Wagner, G, Nagy, H.M, Zangger, K, Lindermuth, H, Oberer, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Cgi-58 Motif Provides the Molecular Basis of Lipid Droplet Anchoring.
J.Biol.Chem., 290, 2015
|
|
5BXV
 
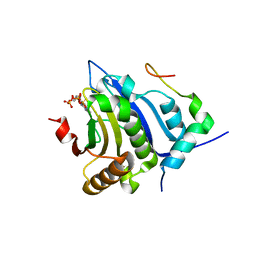 | | eIF4E complex | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | Authors: | Sekiyama, N, Arthanari, H, Papdopoulos, E, Rodriguez-Mias, R.A, Wagner, G, Leger-Abraham, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of the dual activity of 4EGI-1: Dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A3G
 
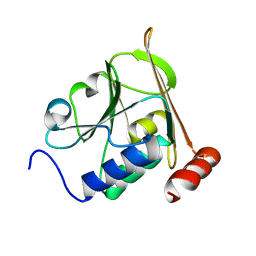 | | Structure of herpesvirus nuclear egress complex subunit M50 | | Descriptor: | M50 | | Authors: | Leigh, K.E, Boeszoermenyi, A, Mansueto, M.S, Sharma, M, Filman, D.J, Coen, D.M, Wagner, G, Hogle, J.M, Arthanari, H. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Herpesvirus Nuclear Egress Complex Subunit Reveals an Interaction Groove that is Essential for Viral Replication
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4TQC
 
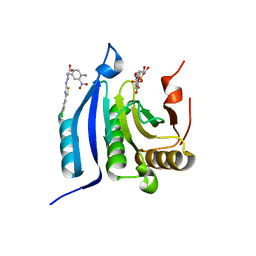 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2S)-3-(4-amino-3-nitrophenyl)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinyl}propanoic acid, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TQB
 
 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2E)-2-{2-[4-(4-bromophenyl)-1,3-thiazol-2-yl]hydrazinylidene}-3-(2-nitrophenyl)propanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TPW
 
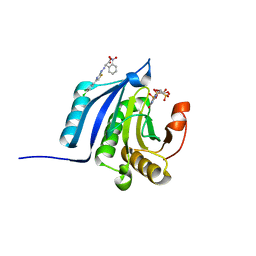 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2E)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinylidene}-3-(2-nitrophenyl)propanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-09 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5GCN
 
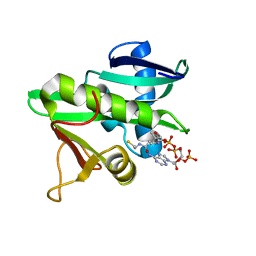 | | CATALYTIC DOMAIN OF TETRAHYMENA GCN5 HISTONE ACETYLTRANSFERASE IN COMPLEX WITH COENZYME A | | Descriptor: | COENZYME A, HISTONE ACETYLTRANSFERASE GCN5 | | Authors: | Lin, Y, Fletcher, C.M, Zhou, J, Allis, C.D, Wagner, G. | | Deposit date: | 1999-03-24 | | Release date: | 1999-07-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A
Nature, 400, 1999
|
|
1I4V
 
 | |
1IIE
 
 | |
1GYF
 
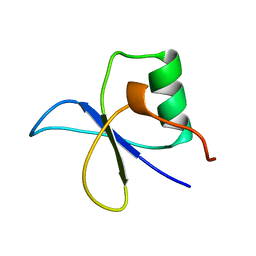 | | GYF DOMAIN FROM HUMAN CD2BP2 PROTEIN | | Descriptor: | PROTEIN (CYTOPLASMIC DOMAIN BINDING PROTEIN (CD2BP2)) | | Authors: | Freund, C, Doetsch, V, Nishizawa, K, Reinherz, E.L, Wagner, G. | | Deposit date: | 1999-04-30 | | Release date: | 2000-01-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences.
Nat.Struct.Biol., 6, 1999
|
|
1J8C
 
 | | Solution Structure of the Ubiquitin-like Domain of hPLIC-2 | | Descriptor: | ubiquitin-like protein hPLIC-2 | | Authors: | Walters, K.J, Kleijnen, M.F, Goh, A.M, Wagner, G, Howley, P.M. | | Deposit date: | 2001-05-21 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a.
Biochemistry, 41, 2002
|
|
1HBW
 
 | | Solution nmr structure of the dimerization domain of the yeast transcriptional activator Gal4 (residues 50-106) | | Descriptor: | REGULATORY PROTEIN GAL4 | | Authors: | Hidalgo, P, Ansari, A.Z, Schmidt, P, Hare, B, Simkovic, N, Farrell, S, Shin, E.J, Ptashne, M, Wagner, G. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Recruitment of the Transcriptional Machinery Through Gal11P: Structure and Interactions of the GAL4 Dimerization Domain
Genes Dev., 15, 2001
|
|
1AJY
 
 | | STRUCTURE AND MOBILITY OF THE PUT3 DIMER: A DNA PINCER, NMR, 13 STRUCTURES | | Descriptor: | PUT3, ZINC ION | | Authors: | Walters, K.J, Dayie, K.T, Reece, R.J, Ptashne, M, Wagner, G. | | Deposit date: | 1997-05-12 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and mobility of the PUT3 dimer.
Nat.Struct.Biol., 4, 1997
|
|
1AJE
 
 | | CDC42 FROM HUMAN, NMR, 20 STRUCTURES | | Descriptor: | CDC42HS | | Authors: | Feltham, J.L, Dotsch, V, Raza, S, Manor, D, Cerione, R.A, Sutcliffe, M.J, Wagner, G, Oswald, R.E. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Definition of the switch surface in the solution structure of Cdc42Hs.
Biochemistry, 36, 1997
|
|
1A66
 
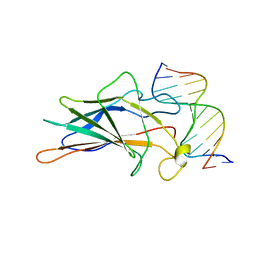 | | SOLUTION NMR STRUCTURE OF THE CORE NFATC1/DNA COMPLEX, 18 STRUCTURES | | Descriptor: | CORE NFATC1, DNA (5'-D(*CP*AP*AP*TP*TP*TP*TP*CP*CP*TP*CP*G)-3'), DNA (5'-D(*CP*GP*AP*GP*GP*AP*AP*AP*AP*TP*TP*G)-3') | | Authors: | Zhou, P, Sun, L.J, Doetsch, V, Wagner, G, Verdine, G.L. | | Deposit date: | 1998-03-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the core NFATC1/DNA complex.
Cell(Cambridge,Mass.), 92, 1998
|
|
1AGT
 
 | | SOLUTION STRUCTURE OF THE POTASSIUM CHANNEL INHIBITOR AGITOXIN 2: CALIPER FOR PROBING CHANNEL GEOMETRY | | Descriptor: | AGITOXIN 2 | | Authors: | Krezel, A.M, Kasibhatla, C, Hidalgo, P, Mackinnon, R, Wagner, G. | | Deposit date: | 1995-04-14 | | Release date: | 1995-07-10 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the potassium channel inhibitor agitoxin 2: caliper for probing channel geometry.
Protein Sci., 4, 1995
|
|
2ME4
 
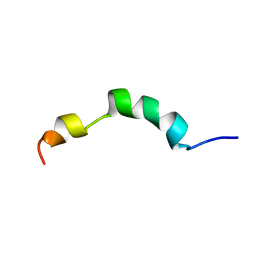 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2ME1
 
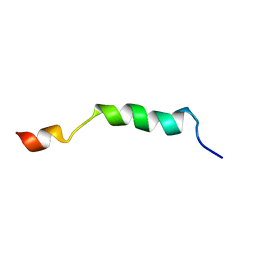 | | HIV-1 gp41 clade B double alanine mutant Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Gp41 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
1IBX
 
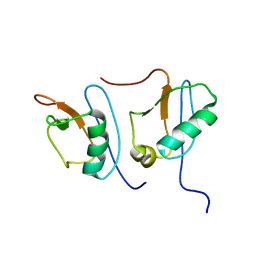 | | NMR STRUCTURE OF DFF40 AND DFF45 N-TERMINAL DOMAIN COMPLEX | | Descriptor: | CHIMERA OF IGG BINDING PROTEIN G AND DNA FRAGMENTATION FACTOR 45, DNA FRAGMENTATION FACTOR 40 | | Authors: | Zhou, P, Lugovskoy, A.A, McCarty, J.S, Li, P, Wagner, G. | | Deposit date: | 2001-03-29 | | Release date: | 2001-05-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1AP8
 
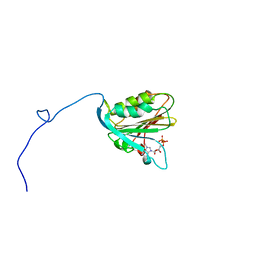 | | TRANSLATION INITIATION FACTOR EIF4E IN COMPLEX WITH M7GDP, NMR, 20 STRUCTURES | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, TRANSLATION INITIATION FACTOR EIF4E | | Authors: | Matsuo, H, Li, H, Mcguire, A.M, Fletcher, M, Gingras, A.C, Sonenberg, N, Wagner, G. | | Deposit date: | 1997-07-25 | | Release date: | 1998-01-28 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein.
Nat.Struct.Biol., 4, 1997
|
|
1AW6
 
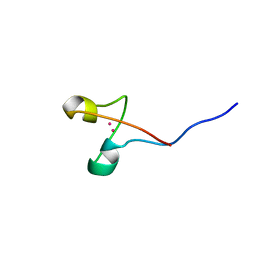 | | GAL4 (CD), NMR, 24 STRUCTURES | | Descriptor: | CADMIUM ION, GAL4 (CD) | | Authors: | Baleja, J.D, Wagner, G. | | Deposit date: | 1997-10-10 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the DNA-binding domain of GAL4 and use of 3J(113Cd,1H) in structure determination.
J.Biomol.NMR, 10, 1997
|
|
2ME3
 
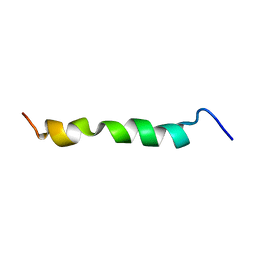 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
1C15
 
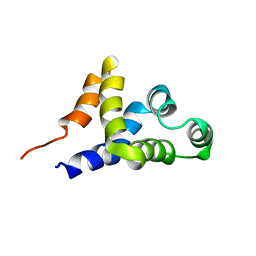 | | SOLUTION STRUCTURE OF APAF-1 CARD | | Descriptor: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1 | | Authors: | Zhou, P, Chou, J, Olea, R.S, Yuan, J, Wagner, G. | | Deposit date: | 1999-07-20 | | Release date: | 1999-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Apaf-1 CARD and its interaction with caspase-9 CARD: a structural basis for specific adaptor/caspase interaction.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1JJS
 
 | | NMR Structure of IBiD, A Domain of CBP/p300 | | Descriptor: | CREB-BINDING PROTEIN | | Authors: | Lin, C.H, Hare, B.J, Wagner, G, Harrison, S.C, Maniatis, T, Fraenkel, E. | | Deposit date: | 2001-07-09 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A small domain of CBP/p300 binds diverse proteins: solution structure and functional studies.
Mol.Cell, 8, 2001
|
|
1JQ4
 
 | | [2Fe-2S] Domain of Methane Monooxygenase Reductase from Methylococcus capsulatus (Bath) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, METHANE MONOOXYGENASE COMPONENT C | | Authors: | Mueller, J, Lugovskoy, A.A, Wagner, G, Lippard, S.J. | | Deposit date: | 2001-08-03 | | Release date: | 2002-01-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the [2Fe-2S] ferredoxin domain from soluble methane monooxygenase reductase and interaction with its hydroxylase.
Biochemistry, 41, 2002
|
|
