2UYP
 
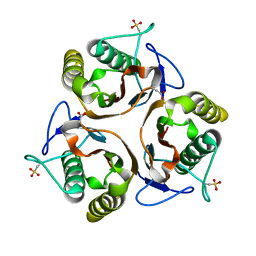 | | Crystal structure of E. coli TdcF with bound propionate | | Descriptor: | PROPANOIC ACID, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
2UYK
 
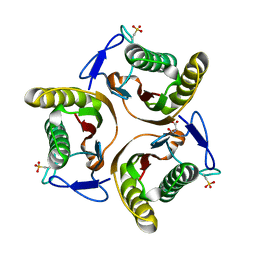 | | Crystal structure of E. coli TdcF with bound serine | | Descriptor: | PROTEIN TDCF, SERINE | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
1H4D
 
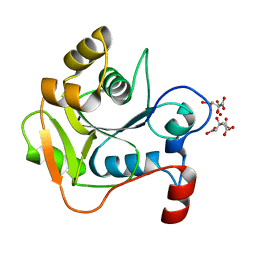 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
1H4E
 
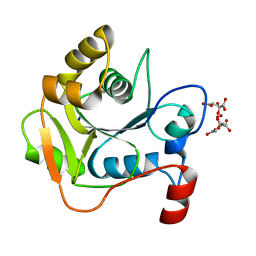 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
1H4C
 
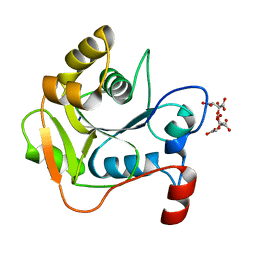 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
1HJL
 
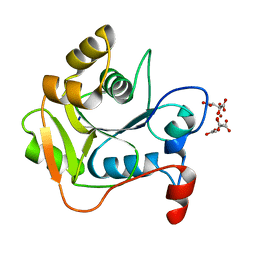 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-27 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
1HJJ
 
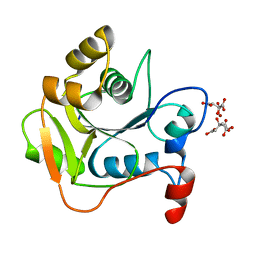 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-27 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
1H9M
 
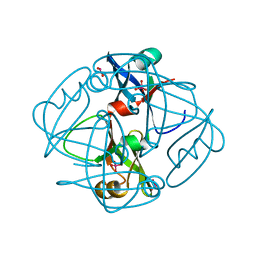 | | Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. PEG-grown form with molybdate bound | | Descriptor: | MOLYBDATE ION, MOLYBDENUM-BINDING-PROTEIN | | Authors: | Delarbre, L, Stevenson, C.E.M, White, D.J, Mitchenall, L.A, Pau, R.N, Lawson, D.M. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Crystal Structures of the Cytoplasmic Molybdate-Binding Protein Modg Suggest a Novel Cooperative Binding Mechanism and Provide Insights Into Ligand-Binding Specificity
J.Mol.Biol., 308, 2001
|
|
1H9J
 
 | | Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. Phosphate-grown form with molybdate and phosphate bound | | Descriptor: | MOLYBDATE ION, MOLYBDENUM-BINDING-PROTEIN, PHOSPHATE ION | | Authors: | Delarbre, L, Stevenson, C.E.M, White, D.J, Mitchenall, L.A, Pau, R.N, Lawson, D.M. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two Crystal Structures of the Cytoplasmic Molybdate-Binding Protein Modg Suggest a Novel Cooperative Binding Mechanism and Provide Insights Into Ligand-Binding Specificity
J.Mol.Biol., 308, 2001
|
|
1H3L
 
 | | N-terminal fragment of SigR from Streptomyces coelicolor | | Descriptor: | RNA POLYMERASE SIGMA FACTOR | | Authors: | Li, W, Stevenson, C.E.M, Burton, N, Jakimowicz, P, Paget, M.S.B, Buttner, M.J, Lawson, D.M, Kleanthous, C. | | Deposit date: | 2002-09-10 | | Release date: | 2002-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.375 Å) | | Cite: | Identification and Structure of the Anti-Sigma Factor-Binding Domain of the Disulfide-Stress Regulated Sigma Factor Sigma(R) from Streptomyces Coelicolor
J.Mol.Biol., 323, 2002
|
|
1H9K
 
 | | Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. Phosphate-grown form with tungstate and phosphate bound | | Descriptor: | MOLYBDENUM-BINDING-PROTEIN, PHOSPHATE ION, TUNGSTATE(VI)ION | | Authors: | Delarbre, L, Stevenson, C.E.M, White, D.J, Mitchenall, L.A, Pau, R.N, Lawson, D.M. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two Crystal Structures of the Cytoplasmic Molybdate-Binding Protein Modg Suggest a Novel Cooperative Binding Mechanism and Provide Insights Into Ligand-Binding Specificity
J.Mol.Biol., 308, 2001
|
|
5NZ7
 
 | | Clostridium thermocellum cellodextrin phosphorylase ligand free form | | Descriptor: | CHLORIDE ION, Cellodextrin phosphorylase | | Authors: | O'Neill, E.C, Pergolizzi, G, Stevenson, C.E.M, Lawson, D.M, Nepogodiev, S.A, Field, R.A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cellodextrin phosphorylase from Ruminiclostridium thermocellum: X-ray crystal structure and substrate specificity analysis.
Carbohydr. Res., 451, 2017
|
|
5NZ8
 
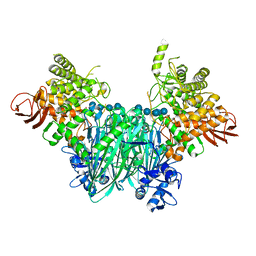 | | Clostridium thermocellum cellodextrin phosphorylase with cellotetraose and phosphate bound | | Descriptor: | Cellodextrin phosphorylase, PHOSPHATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | O'Neill, E.C, Pergolizzi, G, Stevenson, C.E.M, Lawson, D.M, Nepogodiev, S.A, Field, R.A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cellodextrin phosphorylase from Ruminiclostridium thermocellum: X-ray crystal structure and substrate specificity analysis.
Carbohydr. Res., 451, 2017
|
|
5NJA
 
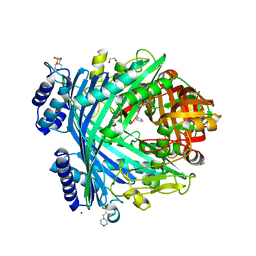 | | E. coli Microcin-processing metalloprotease TldD/E with angiotensin analogue bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HIS-PRO-PHE, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJF
 
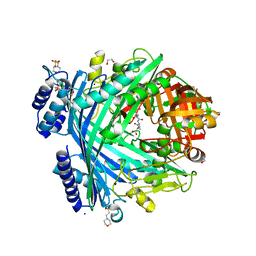 | | E. coli Microcin-processing metalloprotease TldD/E (TldD H262A mutant) with pentapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5O98
 
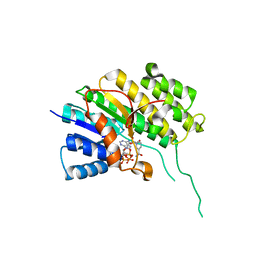 | | Binary complex of Catharanthus roseus Vitrosamine Synthase with NADP+ | | Descriptor: | Alcohol dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Stavrinides, A.K, Tatsis, E.C, Dang, T.T, Caputi, L, Stevenson, C.E.M, Lawson, D.M, Schneider, B, O'Connor, S.E. | | Deposit date: | 2017-06-16 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of a Short-Chain Dehydrogenase from Catharanthus roseus that Produces a New Monoterpene Indole Alkaloid.
Chembiochem, 19, 2018
|
|
5NJB
 
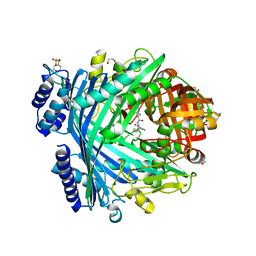 | | E. coli Microcin-processing metalloprotease TldD/E with actinonin bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACTINONIN, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJC
 
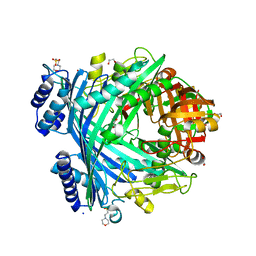 | | E. coli Microcin-processing metalloprotease TldD/E (TldD E263A mutant) with hexapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Metalloprotease PmbA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJ5
 
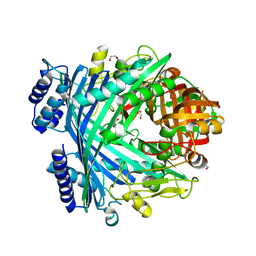 | | E. coli Microcin-processing metalloprotease TldD/E with phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, Metalloprotease PmbA, Metalloprotease TldD, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJ9
 
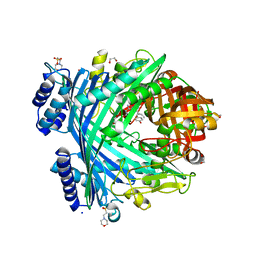 | | E. coli Microcin-processing metalloprotease TldD/E with DRVY angiotensin fragment bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ASP-ARG-VAL-TYR, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
2WMC
 
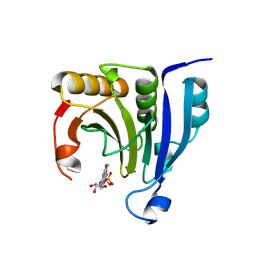 | | Crystal structure of eukaryotic initiation factor 4E from Pisum sativum | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | Authors: | Ashby, J.A, Stevenson, C.E.M, Maule, A.J, Lawson, D.M. | | Deposit date: | 2009-06-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Mutational Analysis of Eif4E in Relation to Sbm1 Resistance to Pea Seed-Borne Mosaic Virus in Pea.
Plos One, 6, 2011
|
|
6F86
 
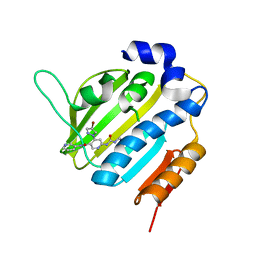 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 4-(4-bromanylpyrazol-1-yl)-6-(ethylcarbamoylamino)-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F94
 
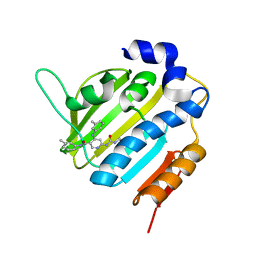 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(3-methyphenyl)amino]-N-(3-methyphenyl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-~{N}-(3-methylphenyl)-4-[(3-methylphenyl)amino]pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F9H
 
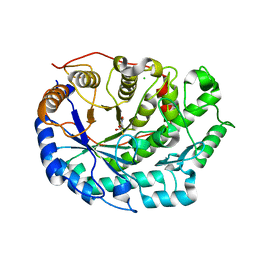 | | Crystal structure of Barley Beta-Amylase complexed with 4-S-alpha-D-glucopyranosyl-(1,4-dideoxy-4-thio-nojirimycin) | | Descriptor: | 1,4-dideoxy-4-thio-nojirimycin, Beta-amylase, CHLORIDE ION, ... | | Authors: | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
6F9L
 
 | | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose | | Descriptor: | Beta-amylase, CHLORIDE ION, alpha-D-glucopyranose-(1-4)-3-deoxy-3-fluoro-alpha-D-glucopyranose | | Authors: | Tantanarat, K, Stevenson, C.E.M, Rejzek, M, Lawson, D.M, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose
To be published
|
|
