8D29
 
 | |
3JXO
 
 | | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima | | Descriptor: | TrkA-N domain protein | | Authors: | Deller, M.C, Johnson, H.A, Miller, M, Spraggon, G, Wilson, I.A, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-09-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima
To be Published
|
|
6BI2
 
 | |
6BHZ
 
 | | Trastuzumab Fab D185A (Light Chain) Mutant. | | Descriptor: | 1,2-ETHANEDIOL, Trastuzumab Anti-HER2 Fab Heavy Chain, Trastuzumab Anti-HER2 Fab Light Chain D185A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Tuning a Protein-Labeling Reaction to Achieve Highly Site Selective Lysine Conjugation.
Chembiochem, 19, 2018
|
|
6BI0
 
 | | Trastuzumab Fab N158A, D185A, K190A (Light Chain) Triple Mutant. | | Descriptor: | 1,2-ETHANEDIOL, Trastuzumab anti-HER2 Fab Heavy Chain, Trastuzumab anti-HER2 Fab Light Chain | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Tuning a Protein-Labeling Reaction to Achieve Highly Site Selective Lysine Conjugation.
Chembiochem, 19, 2018
|
|
4CJD
 
 | | Crystal structure of Neisseria meningitidis trimeric autotransporter and vaccine antigen NadA | | Descriptor: | IODIDE ION, NADA | | Authors: | Malito, E, Biancucci, M, Spraggon, G, Bottomley, M.J. | | Deposit date: | 2013-12-19 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Structure of the Meningococcal Vaccine Antigen Nada and Epitope Mapping of a Bactericidal Antibody.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2QTW
 
 | | The Crystal Structure of PCSK9 at 1.9 Angstroms Resolution Reveals structural homology to Resistin within the C-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Hampton, E.N, Knuth, M.W, Li, J, Harris, J.L, Lesley, S.A, Spraggon, G. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2V9K
 
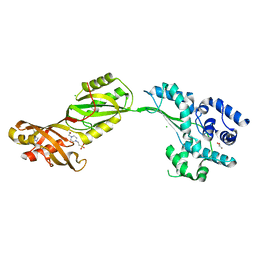 | | Crystal structure of human PUS10, a novel pseudouridine synthase. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | McCleverty, C.J, Hornsby, M, Spraggon, G, Kreusch, A. | | Deposit date: | 2007-08-23 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Pus10, a Novel Pseudouridine Synthase.
J.Mol.Biol., 373, 2007
|
|
2VP8
 
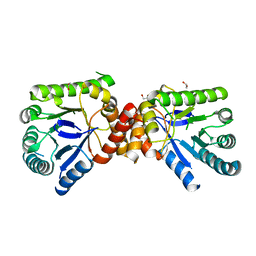 | | Structure of Mycobacterium tuberculosis Rv1207 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPTEROATE SYNTHASE 2 | | Authors: | Gengenbacher, M, Xu, T, Niyomwattanakit, P, Spraggon, G, Dick, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Biochemical and Structural Characterization of the Putative Dihydropteroate Synthase Ortholog Rv1207 of Mycobacterium Tuberculosis.
Fems Microbiol.Lett., 287, 2008
|
|
4BIH
 
 | | Crystal structure of the conserved staphylococcal antigen 1A, Csa1A | | Descriptor: | CALCIUM ION, UNCHARACTERIZED LIPOPROTEIN SAOUHSC_00053 | | Authors: | Malito, E, Bottomley, M.J, Spraggon, G, Schluepen, C, Liberatori, S. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.459 Å) | | Cite: | Mining the Bacterial Unknown Proteome: Identification and Characterization of a Novel Family of Highly Conserved Protective Antigens in Staphylococcus Aureus
Biochem.J., 455, 2013
|
|
4B8Y
 
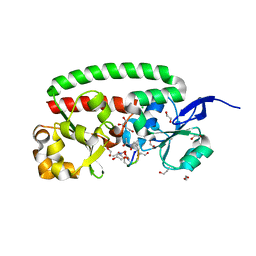 | | Ferrichrome-bound FhuD2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Malito, E, Bottomley, M.J, Spraggon, G. | | Deposit date: | 2012-08-31 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Staphylococcus Aureus Virulence Factor and Vaccine Candidate Fhud2.
Biochem.J., 449, 2013
|
|
3L4B
 
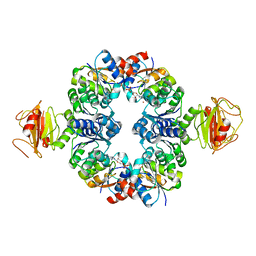 | | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima | | Descriptor: | ADENOSINE MONOPHOSPHATE, TrkA K+ Channel protein TM1088A, TrkA K+ Channel protein TM1088B, ... | | Authors: | Deller, M.C, Johnson, H.A, Miller, M, Spraggon, G, Wilson, I.A, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima
To be Published
|
|
2AQX
 
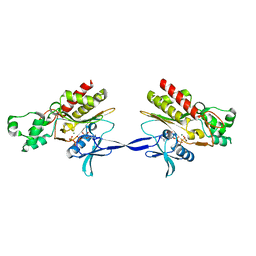 | | Crystal Structure of the Catalytic and CaM-Binding domains of Inositol 1,4,5-Trisphosphate 3-Kinase B | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PREDICTED: inositol 1,4,5-trisphosphate 3-kinase B | | Authors: | Chamberlain, P.P, Sandberg, M.L, Sauer, K, Cooke, M.P, Lesley, S.A, Spraggon, G. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into enzyme regulation for inositol 1,4,5-trisphosphate 3-kinase B
Biochemistry, 44, 2005
|
|
3FCA
 
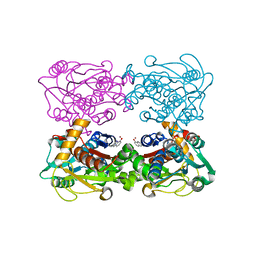 | | Genetic Incorporation of a Metal-ion Chelating Amino Acid into proteins as biophysical probe | | Descriptor: | Cysteine synthase, ZINC ION | | Authors: | Wang, F, Lee, H, Spraggon, G, Schultz, P.G. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Genetic incorporation of a metal-ion chelating amino acid into proteins as a biophysical probe.
J.Am.Chem.Soc., 131, 2009
|
|
4AE1
 
 | |
4AE0
 
 | |
3R5L
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayyar, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-18 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5R
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3QE4
 
 | | An evolved aminoacyl-tRNA Synthetase with atypical polysubstrate specificity | | Descriptor: | 4-cyano-L-phenylalanine, Tyrosyl-tRNA synthetase | | Authors: | Young, D.D, Young, T.S, Jahnz, M, Ahmad, I, Spraggon, G, Schultz, P.G. | | Deposit date: | 2011-01-19 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Evolved Aminoacyl-tRNA Synthetase with Atypical Polysubstrate Specificity .
Biochemistry, 50, 2011
|
|
3R5P
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | Deazaflavin-dependent nitroreductase, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Y
 
 | | Structure of a Deazaflavin-dependent nitroreductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5W
 
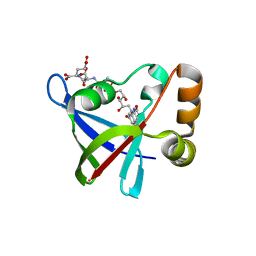 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Z
 
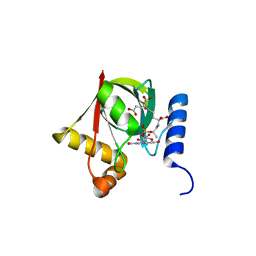 | | Structure of a Deazaflavin-dependent reductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
1FZF
 
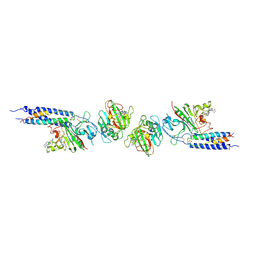 | | CRYSTAL STRUCTURE OF FRAGMENT DOUBLE-D FROM HUMAN FIBRIN WITH THE PEPTIDE LIGAND GLY-HIS-ARG-PRO-AMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Doolittle, R.F. | | Deposit date: | 1998-12-28 | | Release date: | 1999-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Biochemistry, 38, 1999
|
|
1FZE
 
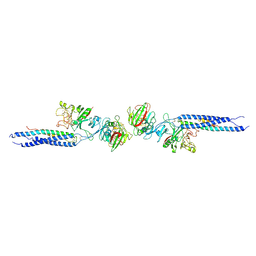 | | CRYSTAL STRUCTURE OF FRAGMENT DOUBLE-D FROM HUMAN FIBRIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Doolittle, R.F. | | Deposit date: | 1998-12-23 | | Release date: | 1999-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro-amide.
Biochemistry, 38, 1999
|
|
