3VV8
 
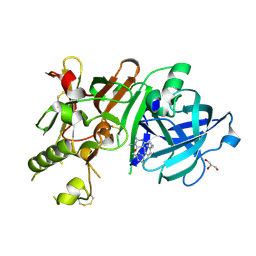 | | Crystal structure of beta secetase in complex with 2-amino-3-methyl-6-((1S,2R)-2-(3'-methylbiphenyl-4-yl)cyclopropyl)pyrimidin-4(3H)-one | | Descriptor: | 2-amino-3-methyl-6-[(1S,2R)-2-(3'-methylbiphenyl-4-yl)cyclopropyl]pyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL | | Authors: | Yonezawa, S, Yamamoto, T, Yamakawa, H, Muto, C, Hosono, M, Hattori, K, Higashino, K, Sakagami, M, Togame, H, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-24 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors: effect of a cyclopropane ring to induce an alternative binding mode.
J.Med.Chem., 55, 2012
|
|
3VV7
 
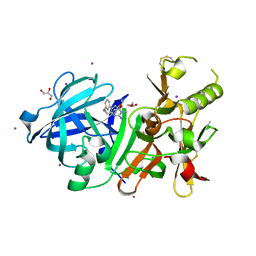 | | Crystal Structure of beta secetase in complex with 2-amino-6-((1S,2R)-2-(3'-methoxybiphenyl-3-yl)cyclopropyl)-3-methylpyrimidin-4(3H)-one | | Descriptor: | 2-amino-6-[(1S,2R)-2-(3'-methoxybiphenyl-3-yl)cyclopropyl]-3-methylpyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL, ... | | Authors: | Yonezawa, S, Yamamoto, T, Yamakawa, H, Muto, C, Hosono, M, Hattori, K, Higashino, K, Sakagami, M, Togame, H, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-24 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors: effect of a cyclopropane ring to induce an alternative binding mode.
J.Med.Chem., 55, 2012
|
|
3VW2
 
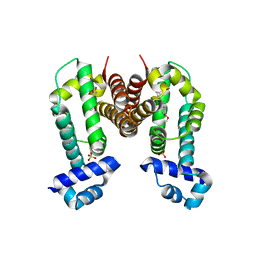 | | Crystal Strucuture of The Berberine-bound Form of RamR (Transcriptional Regurator of TetR Family) from Salmonella Typhimurium | | Descriptor: | BERBERINE, Putative regulatory protein, SULFATE ION | | Authors: | Sakurai, K, Nikaido, E, Nakashima, R, Yamasaki, S, Yamaguchi, A, Nishino, K. | | Deposit date: | 2012-07-30 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The crystal structure of multidrug-resistance regulator RamR with multiple drugs
Nat Commun, 4, 2013
|
|
2MQ9
 
 | |
2K6I
 
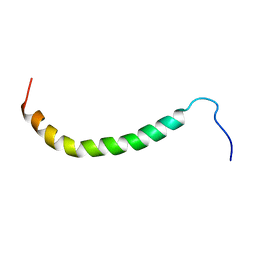 | | The domain features of the peripheral stalk subunit H of the methanogenic A1AO ATP synthase and the NMR solution structure of H1-47 | | Descriptor: | Uncharacterized protein MJ0223 | | Authors: | Biukovic, N, Gayen, S, Pervushin, K, Gruber, G, Biukovic, G. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Domain features of the peripheral stalk subunit H of the methanogenic A1AO ATP synthase and the NMR solution structure of H(1-47).
Biophys.J., 97, 2009
|
|
2LGT
 
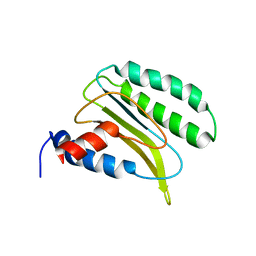 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for QFM(Y)F | | Descriptor: | Eukaryotic peptide chain release factor subunit 1 | | Authors: | Wong, L.E, Li, Y, Pillay, S, Pervushin, K. | | Deposit date: | 2011-08-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Selectivity of stop codon recognition in translation termination is modulated by multiple conformations of GTS loop in eRF1
Nucleic Acids Res., 2012
|
|
3VVZ
 
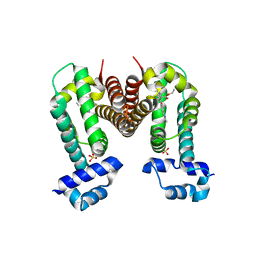 | | Crystal Strucuture of The Rhodamine 6G-Bound Form of RamR (Transcriptional Regurator of TetR Family) from Salmonella Typhimurium | | Descriptor: | Putative regulatory protein, RHODAMINE 6G, SULFATE ION | | Authors: | Sakurai, K, Nikaido, E, Nakashima, R, Yamasaki, S, Yamaguchi, A, Nishino, K. | | Deposit date: | 2012-07-30 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The crystal structure of multidrug-resistance regulator RamR with multiple drugs
Nat Commun, 4, 2013
|
|
2M1C
 
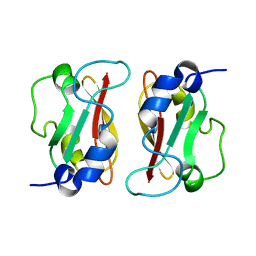 | | HADDOCK structure of GtYybT PAS Homodimer | | Descriptor: | DHH subfamily 1 protein | | Authors: | Liang, Z.X, Pervushin, K, Tan, E, Rao, F, Pasunooti, S, Soehano, I, Lescar, J. | | Deposit date: | 2012-11-25 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the PAS Domain of a Thermophilic YybT Protein Homolog Reveals a Potential Ligand-binding Site.
J.Biol.Chem., 288, 2013
|
|
2MIA
 
 | |
2MI9
 
 | |
4IMN
 
 | | Crystal structure of wild type human Lipocalin PGDS bound with PEG MME 2000 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Lipocalin-type prostaglandin-D synthase | | Authors: | Lim, S.M, Chen, D, Teo, H, Roos, A, Nyman, T, Tresaugues, L, Pervushin, K, Nordlund, P. | | Deposit date: | 2013-01-03 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and dynamic insights into substrate binding and catalysis of human lipocalin prostaglandin D synthase.
J.Lipid Res., 54, 2013
|
|
4IMO
 
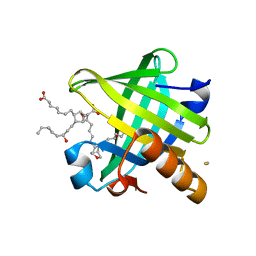 | | Crystal structure of wild type human Lipocalin PGDS in complex with substrate analog U44069 | | Descriptor: | (5E)-7-{(1R,4S,5S,6R)-5-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-6-yl}hept-5-enoic acid, Lipocalin-type prostaglandin-D synthase, THIOCYANATE ION | | Authors: | Lim, S.M, Chen, D, Teo, H, Roos, A, Nyman, T, Tresaugues, L, Pervushin, K, Nordlund, P. | | Deposit date: | 2013-01-03 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and dynamic insights into substrate binding and catalysis of human lipocalin prostaglandin D synthase.
J.Lipid Res., 54, 2013
|
|
5WY9
 
 | |
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
2GTV
 
 | |
3AX8
 
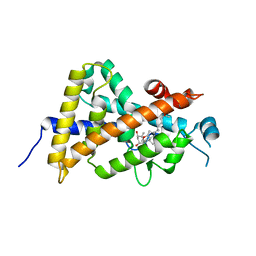 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 15alpha-methoxy-1alpha,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3S,3aS,7aR)-1-[(2R)-6-hydroxy-6-methyl-heptan-2-yl]-3-methoxy-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-03-30 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New C15-substituted active vitamin D3
Org.Lett., 13, 2011
|
|
7TXR
 
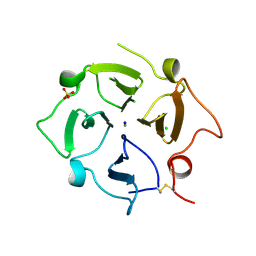 | |
7U68
 
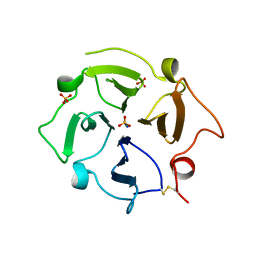 | |
5Z9H
 
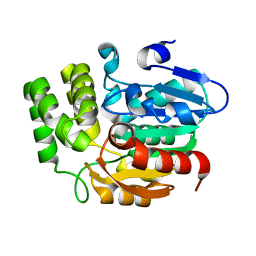 | | Crystal structure of KAI2_ply2(A219V) | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
5Z9G
 
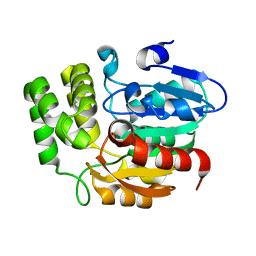 | | Crystal structure of KAI2 | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
5HBC
 
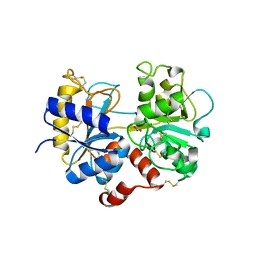 | | Intermediate structure of iron-saturated C-lobe of bovine lactoferrin at 2.79 Angstrom resolution indicates the softening of iron coordination | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, FE (III) ION, ... | | Authors: | Singh, A, Rastogi, N, Singh, P.K, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-12-31 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of iron saturated C-lobe of bovine lactoferrin at pH 6.8 indicates a weakening of iron coordination
Proteins, 84, 2016
|
|
3JRS
 
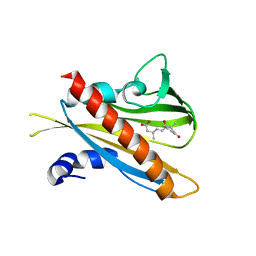 | | Crystal structure of (+)-ABA-bound PYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
3JRQ
 
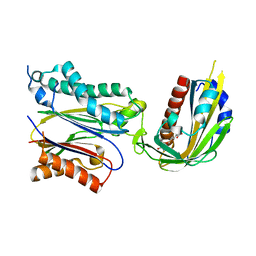 | | Crystal structure of (+)-ABA-bound PYL1 in complex with ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Protein phosphatase 2C 56, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
6Q76
 
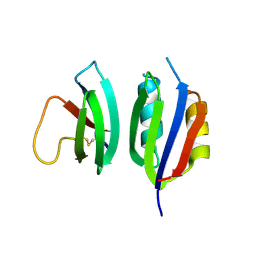 | |
2DDQ
 
 | |
