3GVJ
 
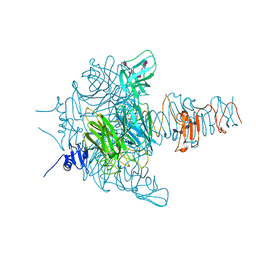 | | Crystal structure of an endo-neuraminidaseNF mutant | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
3GUD
 
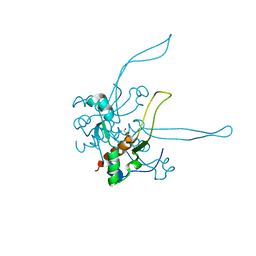 | | Crystal structure of a novel intramolecular chaperon | | Descriptor: | BROMIDE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-29 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3GVL
 
 | | Crystal Structure of endo-neuraminidaseNF | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-beta-neuraminic acid | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
3GVK
 
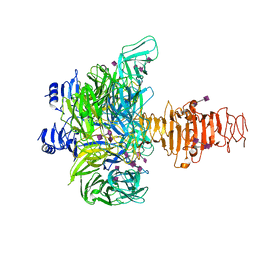 | | Crystal structure of endo-neuraminidase NF mutant | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
3O36
 
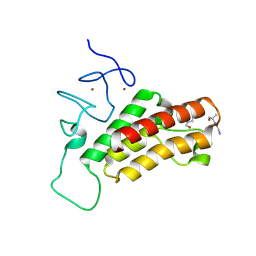 | |
3O35
 
 | |
3O33
 
 | |
3O34
 
 | |
3O37
 
 | |
3PQI
 
 | |
3QR7
 
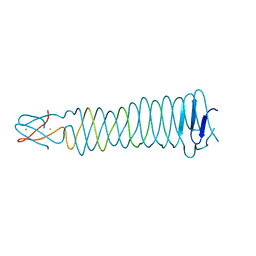 | | Crystal structure of the C-terminal fragment of the bacteriophage P2 membrane-piercing protein gpV | | Descriptor: | Baseplate assembly protein V, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Browning, C, Shneider, M, Leiman, P.G. | | Deposit date: | 2011-02-17 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Phage pierces the host cell membrane with the iron-loaded spike.
Structure, 20, 2012
|
|
3PQH
 
 | |
3QR8
 
 | |
