1W34
 
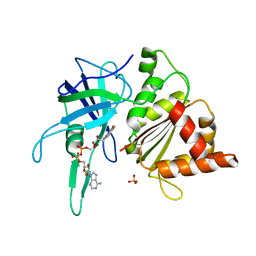 | | FERREDOXIN-NADP REDUCTASE (MUTATION: Y 303 S) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Perez-Dorado, I, Medina, M, Julvez, M.M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2004-07-13 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
3OBA
 
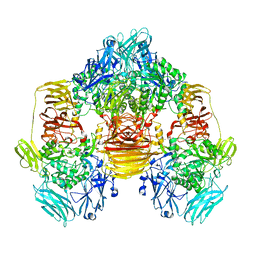 | | Structure of the beta-galactosidase from Kluyveromyces lactis | | Descriptor: | Beta-galactosidase, GLYCEROL, MANGANESE (III) ION | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Becerra, M, Gonzalez-Siso, I, Cerdan, M.E, Sanz-Aparicio, J. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of specificity in tetrameric Kluyveromyces lactis beta-galactosidase.
J.Struct.Biol., 177, 2012
|
|
6EPB
 
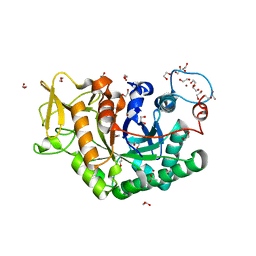 | | Structure of Chitinase 42 from Trichoderma harzianum | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Endochitinase 42, ... | | Authors: | Ramirez-Escudero, M, Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Use of chitin and chitosan to produce new chitooligosaccharides by chitinase Chit42: enzymatic activity and structural basis of protein specificity.
Microb. Cell Fact., 17, 2018
|
|
6EKZ
 
 | |
6EKW
 
 | |
6EKX
 
 | |
6EL4
 
 | |
6EKY
 
 | |
1O90
 
 | | Methionine Adenosyltransferase complexed with a L-methionine analogue | | Descriptor: | (2S,4S)-2-AMINO-4,5-EPOXIPENTANOIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-10 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
2O9P
 
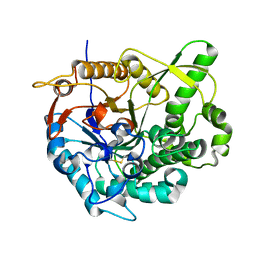 | | beta-glucosidase B from Paenibacillus polymyxa | | Descriptor: | Beta-glucosidase B | | Authors: | Isorna, P, Polaina, J, Sanz-Aparicio, J. | | Deposit date: | 2006-12-14 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
1O92
 
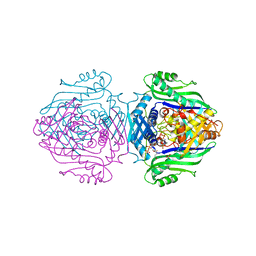 | | Methionine Adenosyltransferase complexed with ADP and a L-methionine analogue | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-2-AMINO-4-METHOXY-CIS-BUT-3-ENOIC ACID, MAGNESIUM ION, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-10 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
1O9T
 
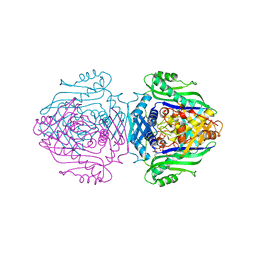 | | Methionine adenosyltransferase complexed with both substrates ATP and methionine | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, METHIONINE, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-18 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
1O93
 
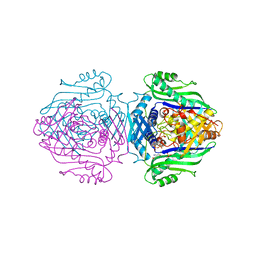 | | Methionine Adenosyltransferase complexed with ATP and a L-methionine analogue | | Descriptor: | (2S,4S)-2-AMINO-4,5-EPOXIPENTANOIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-10 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
2O9R
 
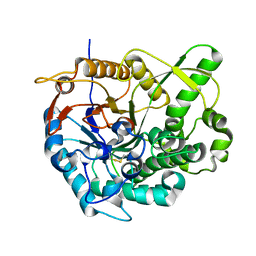 | | beta-glucosidase B complexed with thiocellobiose | | Descriptor: | Beta-glucosidase B, beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose | | Authors: | Isorna, P, Polaina, J, Sanz-Aparicio, J. | | Deposit date: | 2006-12-14 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
2O9T
 
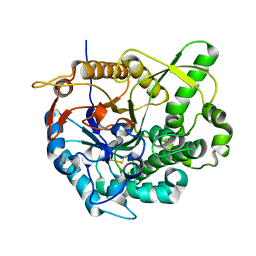 | |
5K6N
 
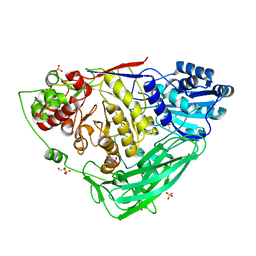 | |
5K6O
 
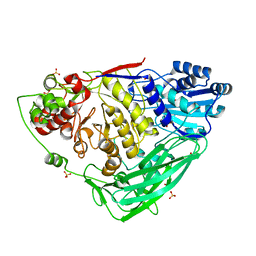 | |
5K6M
 
 | |
5K6L
 
 | |
7ZR3
 
 | |
1H85
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH VAL 136 REPLACED BY LEU (V136L) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a Cluster of Hydrophobic Residues Near the Fad Cofactor in Anabaena Pcc 7119 Ferredoxin-Nadp+ Reductase for Optimal Complex Formation and Electron Transfer to Ferredoxin
J.Biol.Chem., 276, 2001
|
|
3LRL
 
 | | Structure of alfa-galactosidase (MEL1) from Saccharomyces cerevisiae with melibiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
3LRK
 
 | | Structure of alfa-galactosidase (MEL1) from Saccharomyces cerevisiae | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
3LRM
 
 | | Structure of alfa-galactosidase from Saccharomyces cerevisiae with raffinose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
5JUV
 
 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-b-Galactopyranosyl galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-05-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
