8RUA
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV C135A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
8RUD
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV K138A mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
8RUG
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV C189A mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
7P8E
 
 | | Crystal structure of the Receiver domain of M. truncatula cytokinin receptor MtCRE1 | | Descriptor: | CALCIUM ION, Receiver domain of histidine kinase | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
7P8D
 
 | | Crystal structure of the Receiver domain of A. thaliana cytokinin receptor AtCRE1 in complex with Mg2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
7P8C
 
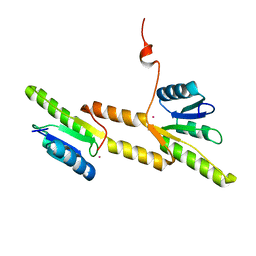 | | Crystal structure of the Receiver domain of A. thaliana cytokinin receptor AtCRE1 in complex with K+ | | Descriptor: | POTASSIUM ION, Receiver domain of histidine kinase 4 | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
8AQL
 
 | | High-resolution crystal structure of human SHMT2 | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine hydroxymethyltransferase, ... | | Authors: | Tran, L.H, Ruszkowski, M. | | Deposit date: | 2022-08-12 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | High-resolution crystal structure of human SHMT2
To Be Published
|
|
7NEN
 
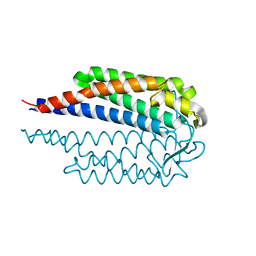 | |
8OWN
 
 | |
8OWM
 
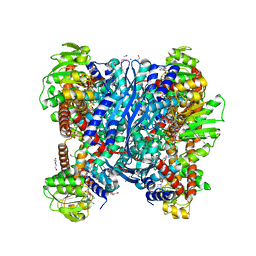 | | Crystal structure of glutamate dehydrogenase 2 from Arabidopsis thaliana binding Ca, NAD and 2,2-dihydroxyglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2,2-bis(oxidanyl)pentanedioic acid, CALCIUM ION, ... | | Authors: | Grzechowiak, M, Ruszkowski, M. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional studies of Arabidopsis thaliana glutamate dehydrogenase isoform 2 demonstrate enzyme dynamics and identify its calcium binding site.
Plant Physiol Biochem., 201, 2023
|
|
7BML
 
 | |
6ZTV
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | 1,2-ETHANEDIOL, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
6ZTX
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | 1,2-ETHANEDIOL, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
6CZX
 
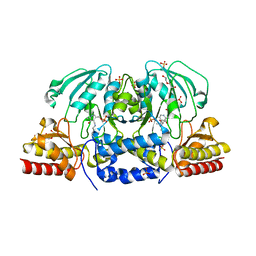 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with PLP internal aldimine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DI(HYDROXYETHYL)ETHER, Phosphoserine aminotransferase 1, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
6CZZ
 
 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with PLP-phosphoserine geminal diamine intermediate | | Descriptor: | PHOSPHOSERINE, PYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase 1, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
8QI7
 
 | |
6DZG
 
 | |
8QAW
 
 | | Medicago truncatula HISN5 (IGPD) in complex with MN, IMD, EDO, FMT, GOL and TRS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
8QAX
 
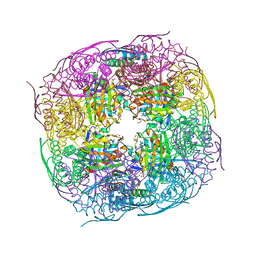 | | Medicago truncatula HISN5 (IGPD) in complex with MN, FMT, GOL and TRS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
8QAY
 
 | | Medicago truncatula HISN5 (IGPD) in complex with MN, FMT, ACT, CIT, EDO, SO4 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CITRIC ACID, ... | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
8QAV
 
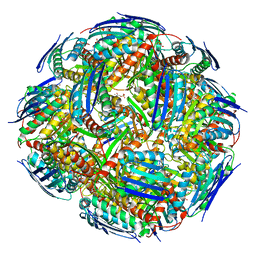 | | Medicago truncatula HISN5 (IGPD) in complex with MN and IG2 | | Descriptor: | (2S,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
6SVS
 
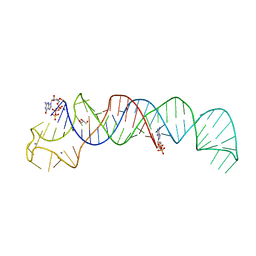 | | Crystal Structure of U:A-U-rich RNA triple helix with 11 consecutive base triples | | Descriptor: | ADENOSINE-5'-PHOSPHATE-2',3'-CYCLIC PHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Ruszkowska, A, Ruszkowski, M, Hulewicz, J.P, Dauter, Z, Brown, J.A. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular structure of a U•A-U-rich RNA triple helix with 11 consecutive base triples.
Nucleic Acids Res., 48, 2020
|
|
8BJ1
 
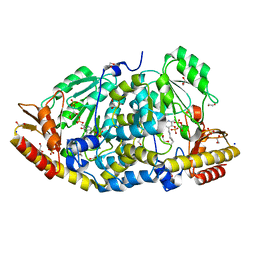 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in the open state | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
8BJ2
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in the closed state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, SODIUM ION, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
8BJ3
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in complex with histidinol-phosphate | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, [(2~{S})-3-(1~{H}-imidazol-4-yl)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]propyl] dihydrogen phosphate, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
