6PUG
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-2`OH-Ethyl-5-OP-U | | Descriptor: | 6-[(2-hydroxyethyl)amino]-5-[(E)-(2-oxopropylidene)amino]pyrimidine-2,4(1H,3H)-dione, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
6PUD
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-5'OH-Pentyl-5-OP-U | | Descriptor: | 6-[(5-hydroxypentyl)amino]-5-[(E)-propylideneamino]pyrimidine-2,4(1H,3H)-dione, Beta-2-microglobulin, Human TCR alpha chain, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
8SHI
 
 | | Valpha3S1 Vbeta13S1 HLA C 0602 VRSRRCLRL | | Descriptor: | Beta-2-microglobulin, MHC class I antigen (Fragment), T cell receptor alpha, ... | | Authors: | Littler, D.R, Anand, S, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.90001726 Å) | | Cite: | Complimentary electrostatics dominate T-cell receptor binding to a psoriasis-associated peptide antigen presented by human leukocyte antigen C∗06:02.
J.Biol.Chem., 299, 2023
|
|
3CKJ
 
 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CITRIC ACID, PHOSPHATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-15 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
8F2R
 
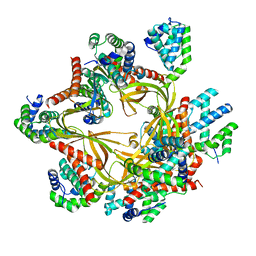 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
8F2U
 
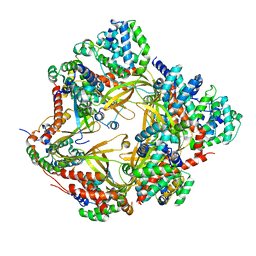 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
3CKO
 
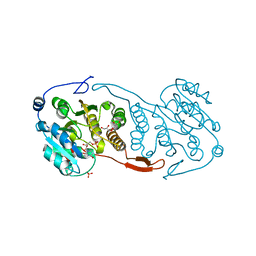 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-16 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
4UU2
 
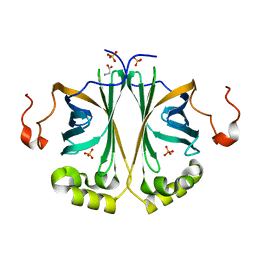 | | Ferulic acid decarboxylase from Enterobacter sp., single mutant | | Descriptor: | FERULIC ACID DECARBOXYLASE, GLYCINE, PHOSPHATE ION, ... | | Authors: | Hromic, A, Pavkov-Keller, T, Steinkellner, G, Lyskowski, A, Wuensch, C, Gross, J, Fuchs, M, Fauland, K, Glueck, S.M, Faber, K, Gruber, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Regioselective Enzymatic Beta-Carboxylation of Para-Hydroxy-Styrene Derivatives Catalyzed by Phenolic Acid Decarboxylases.
Adv. Synth. Catal., 357, 2015
|
|
6PX6
 
 | | HLA-TCR complex | | Descriptor: | DQ2.2-glut-L1, HLA class II histocompatibility antigen DQ alpha chain, HLA class II histocompatibility antigen DQ beta chain, ... | | Authors: | Ting, Y.T, Reid, H.H, Rossjohn, J. | | Deposit date: | 2019-07-24 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.00000334 Å) | | Cite: | A molecular basis for the T cell response in HLA-DQ2.2 mediated celiac disease.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ZQK
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
1SNG
 
 | | Structure of a Thermophilic Serpin in the Native State | | Descriptor: | COG4826: Serine protease inhibitor, SULFATE ION | | Authors: | Fulton, K.F, Buckle, A.M, Cabrita, L.D, Irving, J.A, Butcher, R.E, Smith, I, Reeve, S, Lesk, A.M, Bottomley, S.P, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2004-03-10 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The high resolution crystal structure of a native thermostable serpin reveals the complex mechanism underpinning the stressed to relaxed transition.
J.Biol.Chem., 280, 2005
|
|
4U6Y
 
 | | Crystal Structure of HLA-A*0201 in complex with FLNDK, a 15 mer self-peptide | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gras, S, Chabrol, E, Rossjohn, J. | | Deposit date: | 2014-07-30 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | Naturally Processed Non-canonical HLA-A*02:01 Presented Peptides.
J.Biol.Chem., 290, 2015
|
|
5HHQ
 
 | | Crystal Structure of HLA-A*0201 in complex with M1-L3W | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Gras, S, Josephs, T.M, Rossjohn, J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HHP
 
 | | Crystal Structure of HLA-A*0201 in complex with M1-G4E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Gras, S, Josephs, T.M, Rossjohn, J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4U6X
 
 | | Crystal Structure of HLA-A*0201 in complex with ALQDA, a 15 mer self-peptide | | Descriptor: | ALQDA peptide, ALQDAGDSSRKEYFI, Beta-2-microglobulin, ... | | Authors: | Gras, S, Chabrol, E, Rossjohn, J. | | Deposit date: | 2014-07-30 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Naturally Processed Non-canonical HLA-A*02:01 Presented Peptides.
J.Biol.Chem., 290, 2015
|
|
5HHM
 
 | | Crystal Structure of the JM22 TCR in complex with HLA-A*0201 in complex with M1-F5L | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Gras, S, Josephs, T.M, Rossjohn, J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4UU3
 
 | | Ferulic acid decarboxylase from Enterobacter sp. | | Descriptor: | FERULIC ACID DECARBOXYLASE | | Authors: | Hromic, A, Pavkov-Keller, T, Steinkellner, G, Lyskowski, A, Wuensch, C, Gross, J, Fuchs, M, Fauland, K, Glueck, S.M, Faber, K, Gruber, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Regioselective Enzymatic Beta-Carboxylation of Para-Hydroxy-Styrene Derivatives Catalyzed by Phenolic Acid Decarboxylases.
Adv. Synth. Catal., 357, 2015
|
|
7ZQ4
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ and the oxidated catalytic cysteine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase A, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7ZQ3
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
6PVC
 
 | |
6PVD
 
 | |
1D5S
 
 | | CRYSTAL STRUCTURE OF CLEAVED ANTITRYPSIN POLYMER | | Descriptor: | P1-ARG ANTITRYPSIN | | Authors: | Dunstone, M.A, Dai, W, Whisstock, J.C, Rossjohn, J, Pike, R.N, Feil, S.C, Le Bonneic, B.F, Parker, M.W, Bottomley, S.P. | | Deposit date: | 1999-10-11 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cleaved antitrypsin polymers at atomic resolution.
Protein Sci., 9, 2000
|
|
6OVN
 
 | | Crystal structure of the unliganded Clone 2 TCR | | Descriptor: | Alpha chain Clone 2 TCR, Beta chain Clone 2 TCR, CHLORIDE ION, ... | | Authors: | Shahine, A, Rossjohn, J. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A TCR beta-Chain Motif Biases toward Recognition of Human CD1 Proteins.
J Immunol., 203, 2019
|
|
6OVO
 
 | | Crystal structure of the unliganded PG10 TCR | | Descriptor: | 1,2-ETHANEDIOL, Alpha Chain T-Cell Receptor PG10, Beta Chain T-Cell Receptor PG10, ... | | Authors: | Shahine, A, Rossjohn, J. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A TCR beta-Chain Motif Biases toward Recognition of Human CD1 Proteins.
J Immunol., 203, 2019
|
|
6PUF
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-5'D-5-OP-RU | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
