5M33
 
 | | Structural tuning of CD81LEL (space group P21) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
5M3T
 
 | | Structural tuning of CD81LEL (space group P64) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, CHLORIDE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-17 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
5M2C
 
 | | Structural tuning of CD81LEL (space group P32 1 2) | | Descriptor: | CD81 antigen, PHOSPHATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
5N35
 
 | | Gadolinium phased PBP2 (SSO6202) at 2.2 Ang | | Descriptor: | GADOLINIUM ATOM, GLYCEROL, NITRATE ION, ... | | Authors: | Yan, J, Beattie, T.R, Rojas, A.L, Schermerhorn, K, Gristwood, T, Trinidad, J.C, Albers, S.V, Roversi, P, Gardner, A.F, Abrescia, N.G.A, Bell, S.D. | | Deposit date: | 2017-02-08 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Identification and characterization of a heterotrimeric archaeal DNA polymerase holoenzyme.
Nat Commun, 8, 2017
|
|
5M7F
 
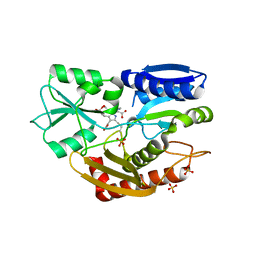 | | Human porphobilinogen deaminase in complex with DPM cofactor | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase, SULFATE ION | | Authors: | Pluta, P, Millet, O, Roversi, P, Rojas, A.L, Gu, S. | | Deposit date: | 2016-10-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural basis of pyrrole polymerization in human porphobilinogen deaminase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5M6R
 
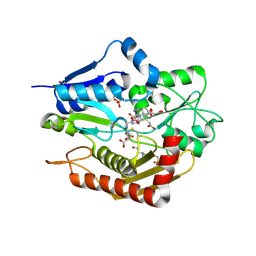 | | Human porphobilinogen deaminase in complex with reaction intermediate | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PHOSPHATE ION, ... | | Authors: | Pluta, P, Millet, O, Roversi, P, Rojas, A.L, Gu, S. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural basis of pyrrole polymerization in human porphobilinogen deaminase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5MTO
 
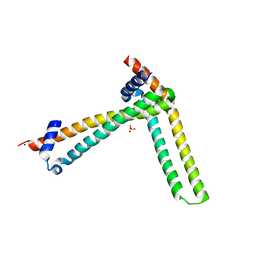 | | N-terminal domain of the human tumor suppressor ING5 C19S mutant | | Descriptor: | Inhibitor of growth protein 5, SODIUM ION, SULFATE ION | | Authors: | Ormaza, G, Buitrago, J.A.R, Roversi, P, Rojas, A.L, Blanco, F.J. | | Deposit date: | 2017-01-10 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Tumor Suppressor ING5 Is a Dimeric, Bivalent Recognition Molecule of the Histone H3K4me3 Mark.
J.Mol.Biol., 431, 2019
|
|
5OSI
 
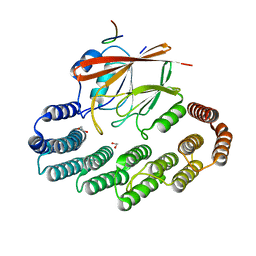 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL harpin loop (163-176) | | Descriptor: | 1,2-ETHANEDIOL, Interaptin, SODIUM ION, ... | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MIY
 
 | | Crystal structure of the E3 ubiquitin ligase RavN from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin ligase RavN, SODIUM ION, ... | | Authors: | Lucas, M, Abascal-Palacios, G, Rojas, A.L, Hierro, A. | | Deposit date: | 2016-11-29 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | RavN is a member of a previously unrecognized group of Legionella pneumophila E3 ubiquitin ligases.
PLoS Pathog., 14, 2018
|
|
5OSH
 
 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL N-terminal domain (1-236) | | Descriptor: | Interaptin, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2GO2
 
 | |
