4JUI
 
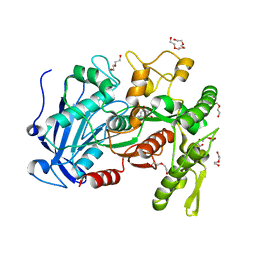 | | crystal structure of tannase from from Lactobacillus plantarum | | Descriptor: | DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, McKinstry, W.J, Chen, Q. | | Deposit date: | 2013-03-24 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7VIT
 
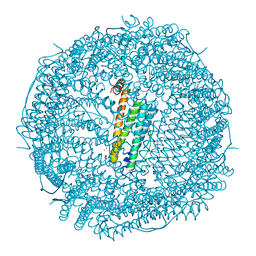 | | Crystal structure of Au(400EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIR
 
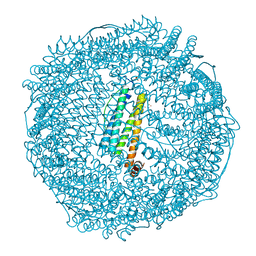 | | Crystal structure of Au(100EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | CADMIUM ION, Ferritin light chain, GOLD ION, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIO
 
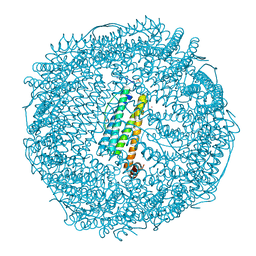 | | Crystal structure of recombinant horse spleen apo-R168H/L169C-Fr | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Wu, J, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIP
 
 | | Crystal structure of Au(10EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIU
 
 | | Crystal structure of Au(200EQ)-apo-R168C/L169C-rHLFr | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIQ
 
 | | Crystal structure of Au(50EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIS
 
 | | Crystal structure of Au(200EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
6X59
 
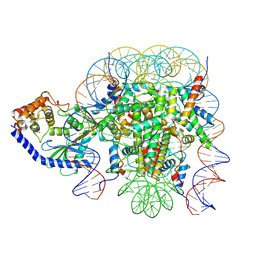 | | The mouse cGAS catalytic domain binding to human assembled nucleosome | | Descriptor: | Cyclic GMP-AMP synthase, DNA, Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
6X5A
 
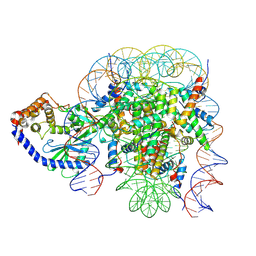 | | The mouse cGAS catalytic domain binding to human nucleosome that purified from HEK293T cells | | Descriptor: | Cyclic GMP-AMP synthase, DNA (natural), Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
1UEF
 
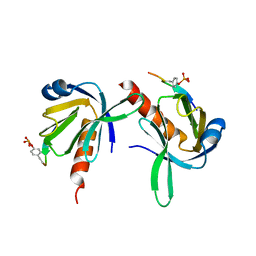 | | Crystal Structure of Dok1 PTB Domain Complex | | Descriptor: | 13-mer peptide from Proto-oncogene tyrosine-protein kinase receptor ret, Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yan, J, Rao, Z. | | Deposit date: | 2003-05-14 | | Release date: | 2004-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.Biol.Chem., 279, 2004
|
|
3G36
 
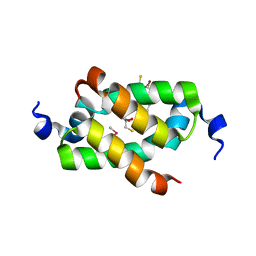 | | Crystal structure of the human DPY-30-like C-terminal domain | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Wang, X, Lou, Z, Bartlam, M, Rao, Z. | | Deposit date: | 2009-02-02 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the C-terminal domain of human DPY-30-like protein: A component of the histone methyltransferase complex
J.Mol.Biol., 390, 2009
|
|
9LCR
 
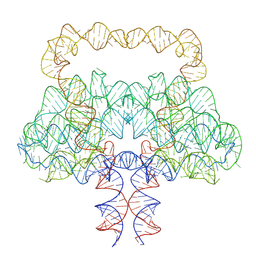 | | Clostridium botulinum OLE RNA dimer | | Descriptor: | MAGNESIUM ION, RNA (578-MER) | | Authors: | Jia, X.Y, Wang, L, Su, Z.M. | | Deposit date: | 2025-01-05 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM reveals mechanisms of natural RNA multivalency.
Science, 388, 2025
|
|
6SMB
 
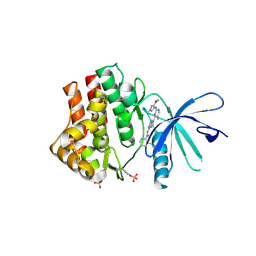 | | Human jak1 kinase domain in complex with inhibitor | | Descriptor: | Tyrosine-protein kinase JAK1, ~{N}-[3-[2-[(3-methoxy-1-methyl-pyrazol-4-yl)amino]-5-methyl-pyrimidin-4-yl]-1~{H}-indol-7-yl]-2-methyl-pyridine-3-carboxamide | | Authors: | Read, J.A, Steuber, H. | | Deposit date: | 2019-08-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery of (2R)-N-[3-[2-[(3-Methoxy-1-methyl-pyrazol-4-yl)amino]pyrimidin-4-yl]-1H-indol-7-yl]-2-(4-methylpiperazin-1-yl)propenamide (AZD4205) as a Potent and Selective Janus Kinase 1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6SM8
 
 | | Human jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-6-[(3~{S})-3-[(1~{S})-2-cyano-1-[4-(7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)pyrazol-1-yl]ethyl]pyrrolidin-1-yl]benzenecarbonitrile, Tyrosine-protein kinase JAK1 | | Authors: | Read, J.A, Steuber, H. | | Deposit date: | 2019-08-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of (2R)-N-[3-[2-[(3-Methoxy-1-methyl-pyrazol-4-yl)amino]pyrimidin-4-yl]-1H-indol-7-yl]-2-(4-methylpiperazin-1-yl)propenamide (AZD4205) as a Potent and Selective Janus Kinase 1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
1P5T
 
 | | Crystal Structure of Dok1 PTB Domain | | Descriptor: | Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yuan, J, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.BIOL.CHEM., 279, 2004
|
|
5EOB
 
 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | 6-[bis(fluoranyl)-[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]quinoline, Hepatocyte growth factor receptor | | Authors: | Liu, Q, Chen, T, Xu, Y. | | Deposit date: | 2015-11-10 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of 6-(difluoro(6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl)methyl)quinoline as a highly potent and selective c-Met inhibitor
Eur.J.Med.Chem., 116, 2016
|
|
6THH
 
 | |
2FQD
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2X7I
 
 | | Crystal structure of mevalonate kinase from methicillin-resistant Staphylococcus aureus MRSA252 | | Descriptor: | CHLORIDE ION, CITRIC ACID, MEVALONATE KINASE | | Authors: | Oke, M, Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-27 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2FQE
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQG
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQF
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
