8VIU
 
 | |
8UTG
 
 | | An RNA G-Quadruplex from the NS5 gene in the West Nile Virus Genome | | Descriptor: | AMMONIUM ION, POTASSIUM ION, RNA (5'-R(*UP*AP*UP*GP*GP*AP*AP*GP*AP*GP*GP*CP*GP*GP*UP*UP*GP*GP*UP*AP*U)-3') | | Authors: | Terrell, J.R, Le, T.T, Wilson, W.D, Siemer, J.L. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of an RNA G-quadruplex from the West Nile virus genome.
Nat Commun, 15, 2024
|
|
8RPH
 
 | | JanthE from Janthinobacterium sp. HH01,ketobutyryl-ThDP | | Descriptor: | (2~{S})-2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-2-yl]-2-oxidanyl-butanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8T4N
 
 | |
8RPJ
 
 | | JanthE from Janthinobacterium sp. HH01 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RPI
 
 | | JanthE from Janthinobacterium sp. HH01, lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8F2Y
 
 | |
8Q2E
 
 | | The 1.68-A X-ray crystal structure of Sporosarcina pasteurii urease inhibited by thiram and bound to dimethylditiocarbamate | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2023-08-02 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Kinetic and structural details of urease inactivation by thiuram disulphides.
J.Inorg.Biochem., 250, 2023
|
|
6NZ2
 
 | |
3DYA
 
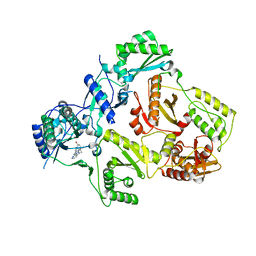 | | HIV-1 RT with non-nucleoside inhibitor annulated Pyrazole 1 | | Descriptor: | 3-[6-bromo-2-fluoro-3-(1H-pyrazolo[3,4-c]pyridazin-3-ylmethyl)phenoxy]-5-chlorobenzonitrile, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, p51 RT | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of annulated pyrazoles as inhibitors of HIV-1 reverse transcriptase
J.Med.Chem., 51, 2008
|
|
3E01
 
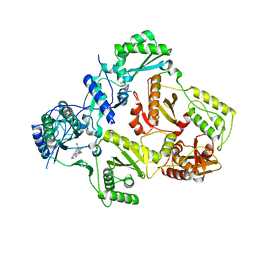 | | HIV-RT with non-nucleoside inhibitor annulated pyrazole 2 | | Descriptor: | 3-[2-bromo-4-(1H-pyrazolo[3,4-c]pyridazin-3-ylmethyl)phenoxy]-5-methylbenzonitrile, Gag-Pol polyprotein | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2008-07-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Design of annulated pyrazoles as inhibitors of HIV-1 reverse transcriptase
J.Med.Chem., 51, 2008
|
|
8EC1
 
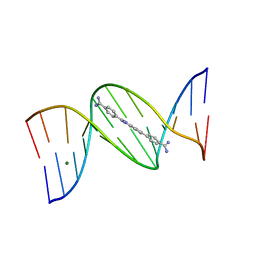 | |
8ED6
 
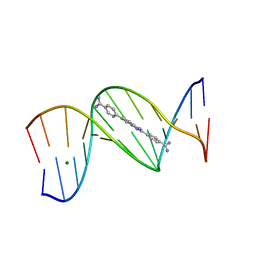 | |
8EDA
 
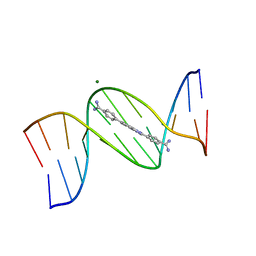 | | An alternating AT dodecamer benzimidazole (DB1476) complex | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-09-03 | | Release date: | 2023-02-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8F2W
 
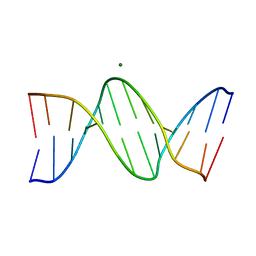 | | Structure of a B-Form Dodecamer: 5'-CGCGAATTCGCG-3 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-08 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8EDB
 
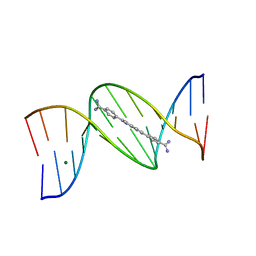 | | 5'-CGCGAATTCGCG-3' and an AT-specific binder (DB1884) complex | | Descriptor: | 4,4'-(1H-indole-2,6-diyl)dibenzenecarboximidamide, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-09-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8F20
 
 | | Structure of an A-tract B-DNA Dodecamer: 5'-CGCAAAAAAGCG-3 | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8FDP
 
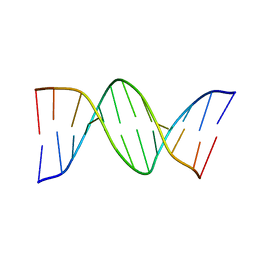 | |
8F94
 
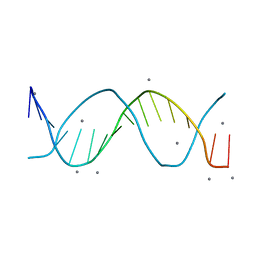 | |
8FB4
 
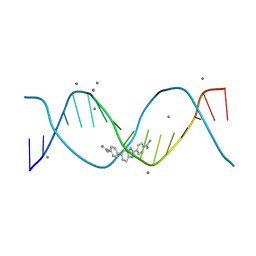 | | Structure of an alternating AT 16-mer bound by diamidine DB1476: 5'-GCTGGATATATCCAGC-3 | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*GP*AP*TP*AP*TP*AP*TP*CP*CP*AP*GP*C)-3') | | Authors: | Terrell, J.R, Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-29 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8FDQ
 
 | |
7KU4
 
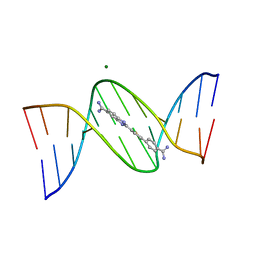 | |
7KWK
 
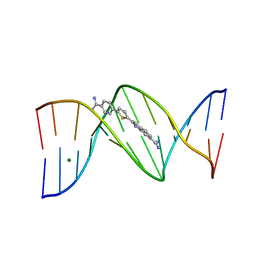 | |
6THL
 
 | | Crystal structure of the complex between RTT106 and BCD1 | | Descriptor: | Box C/D snoRNA protein 1, Rtt106p | | Authors: | Charron, C, Bragantini, B, Manival, X, Charpentier, B. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The box C/D snoRNP assembly factor Bcd1 interacts with the histone chaperone Rtt106 and controls its transcription dependent activity.
Nat Commun, 12, 2021
|
|
3LAL
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with N1-ethyl pyrimidinedione non-nucleoside inhibitor | | Descriptor: | 3-{[3-ethyl-5-(1-methylethyl)-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl]carbonyl}-5-methylbenzonitrile, HIV Reverse transcriptase, SULFATE ION | | Authors: | Lansdon, E.B, Mitchell, M.L. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | N1-Alkyl pyrimidinediones as non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
