8UCW
 
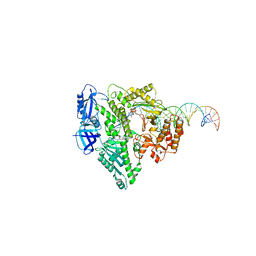 | | Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Chazin, W.C, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UCV
 
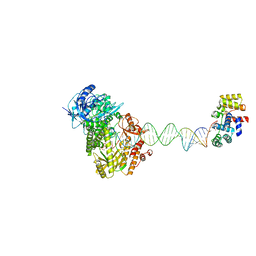 | | Complete DNA termination subcomplex 1 of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Chazin, W.C, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5O
 
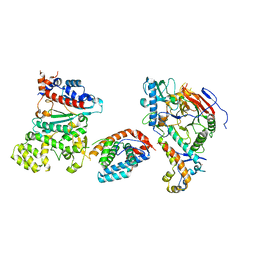 | | Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.99 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5N
 
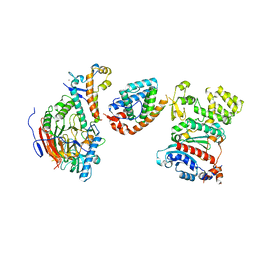 | | Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.56 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5M
 
 | | Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (9.22 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5J3D
 
 | |
3LRV
 
 | |
5ITB
 
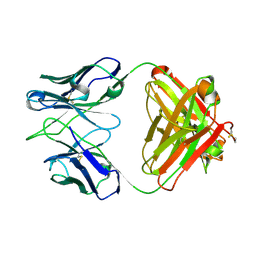 | | Crystal structure of the anti-RSV F Fab 14N4 | | Descriptor: | anti-RSV F Fab 14N4 Heavy chain, anti-RSV F Fab 14N4 Light chain | | Authors: | Mousa, J.J, Crowe, J.E. | | Deposit date: | 2016-03-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for nonneutralizing antibody competition at antigenic site II of the respiratory syncytial virus fusion protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6X58
 
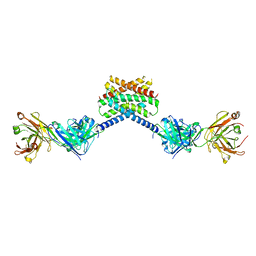 | | MPER-Fluc-Ec2 bound to 10E8v4 antibody | | Descriptor: | 10E8v4 Fab Heavy Chain, 10E8v4 Fab Light Chain, gp41 MPER peptide,Putative fluoride ion transporter CrcB | | Authors: | McIlwain, B.C, Stockbridge, R.B. | | Deposit date: | 2020-05-25 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | N-terminal Transmembrane-Helix Epitope Tag for X-ray Crystallography and Electron Microscopy of Small Membrane Proteins.
J.Mol.Biol., 433, 2021
|
|
6XJ1
 
 | |
6XOB
 
 | |
6XO4
 
 | |
5C1F
 
 | | Structure of the Imp2 F-BAR domain | | Descriptor: | FORMIC ACID, Septation protein imp2 | | Authors: | Vander Kooi, C.W. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3551 Å) | | Cite: | The Tubulation Activity of a Fission Yeast F-BAR Protein Is Dispensable for Its Function in Cytokinesis.
Cell Rep, 14, 2016
|
|
5TV1
 
 | | active arrestin-3 with inositol hexakisphosphate | | Descriptor: | Beta-arrestin-2, GLYCEROL, INOSITOL HEXAKISPHOSPHATE | | Authors: | Chen, Q, Gilbert, N.C, Perry, N.A, Vishniveteskiy, S, Gurevich, V.V, Iverson, T.M. | | Deposit date: | 2016-11-07 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of arrestin-3 activation and signaling.
Nat Commun, 8, 2017
|
|
