6KCA
 
 | |
6KD1
 
 | |
6K1Y
 
 | |
6K1X
 
 | |
6KCD
 
 | |
6K1W
 
 | |
7WBF
 
 | | Crystal structure of lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Nam, K.H. | | Deposit date: | 2021-12-16 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Processing of Multicrystal Diffraction Patterns in Macromolecular Crystallography Using Serial Crystallography Programs.
Crystals, 12, 2022
|
|
7WBD
 
 | |
7WBE
 
 | |
7XF7
 
 | |
7XF6
 
 | | Crystal Structure of Human Lysozyme | | Descriptor: | ACETATE ION, Lysozyme C | | Authors: | Nam, K.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Human Lysozyme Complexed with N-Acetyl-alpha-d-glucosamine.
Appl Sci (Basel), 12, 2022
|
|
7XF8
 
 | |
7DFJ
 
 | |
7DFK
 
 | |
7E25
 
 | | Crystal structure of human FABP7 complexed with palmitic acid | | Descriptor: | Fatty acid-binding protein, brain, GLYCEROL, ... | | Authors: | Nam, K.H. | | Deposit date: | 2021-02-04 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of human brain-type fatty acid-binding protein FABP7 complexed with palmitic acid.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DTF
 
 | |
7E02
 
 | |
7E03
 
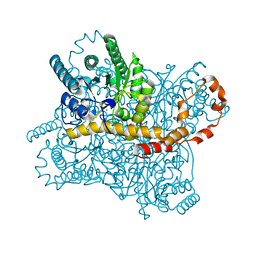 | |
7DTB
 
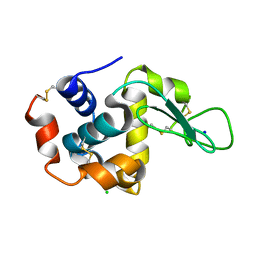 | |
7CJP
 
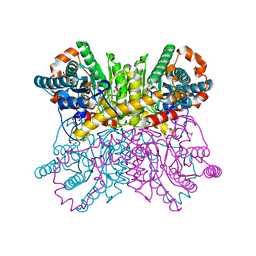 | | Crystal structure of metal-free state of glucose isomerase | | Descriptor: | 1,2-ETHANEDIOL, Xylose isomerase | | Authors: | Nam, K.H. | | Deposit date: | 2020-07-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the metal-free state of glucose isomerase reveals its minimal open configuration for metal binding.
Biochem.Biophys.Res.Commun., 547, 2021
|
|
7CJO
 
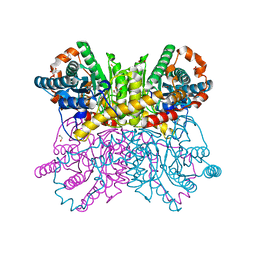 | | Crystal structure of metal-bound state of glucose isomerase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Xylose isomerase | | Authors: | Nam, K.H. | | Deposit date: | 2020-07-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the metal-free state of glucose isomerase reveals its minimal open configuration for metal binding.
Biochem.Biophys.Res.Commun., 547, 2021
|
|
7CJZ
 
 | |
7CK0
 
 | |
6LOF
 
 | | Crystal structure of ZsYellow soaked by Cu2+ | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Nam, K.H. | | Deposit date: | 2020-01-05 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Spectroscopic and Structural Analysis of Cu 2+ -Induced Fluorescence Quenching of ZsYellow.
Biosensors (Basel), 10, 2020
|
|
3II1
 
 | |
