5GZH
 
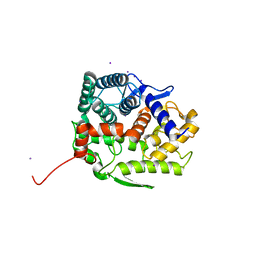 | | Endo-beta-1,2-glucanase from Chitinophaga pinensis - ligand free form | | Descriptor: | Endo-beta-1,2-glucanase, IODIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Abe, K, Nakajima, M, Arakawa, T, Fushinobu, S, Taguchi, H. | | Deposit date: | 2016-09-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family
J. Biol. Chem., 292, 2017
|
|
7ZOI
 
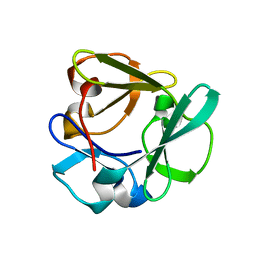 | | Carbohydrate binding domain CBM92-A from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | Descriptor: | Glycoside hydrolase family 18 | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOO
 
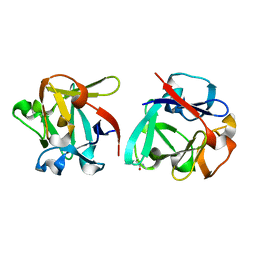 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with gentiobiose | | Descriptor: | Glycoside hydrolase family 18, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZON
 
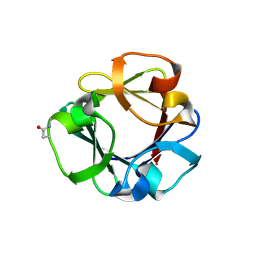 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with glucose | | Descriptor: | Glycoside hydrolase family 18, PENTAETHYLENE GLYCOL, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOH
 
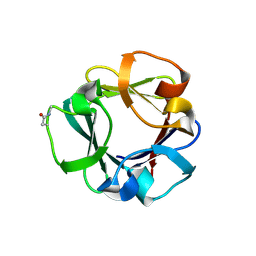 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | Descriptor: | Glycoside hydrolase family 18 | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOP
 
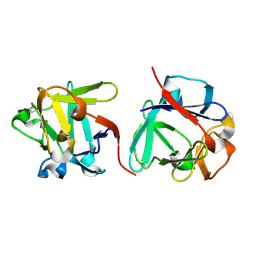 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with sophorose. | | Descriptor: | Glycoside hydrolase family 18, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
5YSD
 
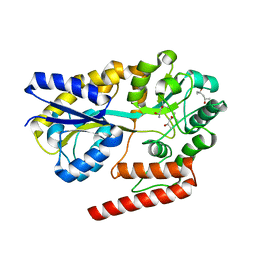 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotriose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSF
 
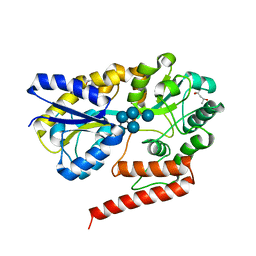 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophoropentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSE
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSB
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in ligand-free form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lin1841 protein, ZINC ION | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
7FE3
 
 | |
7FE4
 
 | |
5B0S
 
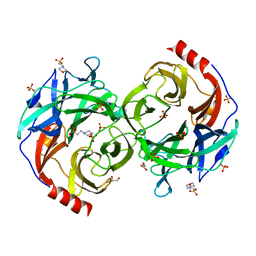 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - beta-1,2-mannotriose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B0R
 
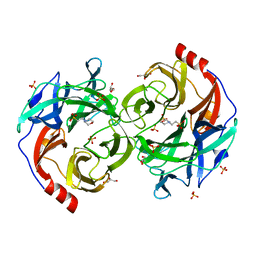 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - beta-1,2-mannobiose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B0P
 
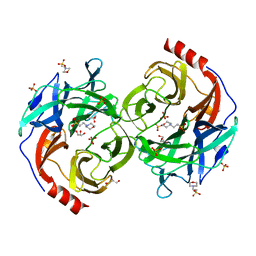 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - glycerol complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B0Q
 
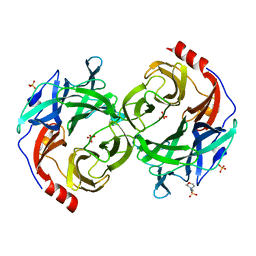 | | beta-1,2-Mannobiose phosphorylase from Listeria innocua - mannose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lin0857 protein, SULFATE ION, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
