1QKB
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KVK | | Descriptor: | ACETATE ION, PEPTIDE LYS-VAL-LYS, PERIPLASMIC OLIGOPEPTIDE-BINDING PROTEIN, ... | | Authors: | Tame, J.R.H, Sleigh, S.H, Wilkinson, A.J. | | Deposit date: | 1999-07-14 | | Release date: | 1999-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and Calorimetric Analysis of Peptide Binding to Oppa Protein
J.Mol.Biol., 291, 1999
|
|
8KDN
 
 | | Structure of LAT1-CD98hc in complex with L-Phe, focused on TMD | | Descriptor: | 4F2 cell-surface antigen heavy chain, Large neutral amino acids transporter small subunit 1, PHENYLALANINE | | Authors: | Lee, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural basis of anticancer drug recognition and amino acid transport by LAT1.
Nat Commun, 16, 2025
|
|
8KDJ
 
 | | Structure of apo inward-open LAT1-CD98h in nanodisc, focused on TMD | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 4F2 cell-surface antigen heavy chain, CHOLESTEROL, ... | | Authors: | Lee, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structural basis of anticancer drug recognition and amino acid transport by LAT1.
Nat Commun, 16, 2025
|
|
8KDH
 
 | | Structure of LAT1-CD98hc in complex with BCH, focused on TMD | | Descriptor: | (1~{S},2~{R},4~{R})-2-azanylbicyclo[2.2.1]heptane-2-carboxylic acid, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural basis of anticancer drug recognition and amino acid transport by LAT1.
Nat Commun, 16, 2025
|
|
8KDD
 
 | | Structure of LAT1-CD98hc-Fab170 in complex with JPH203, consensus map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Fab170 heavy chain, ... | | Authors: | Lee, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structural basis of anticancer drug recognition and amino acid transport by LAT1.
Nat Commun, 16, 2025
|
|
8KDP
 
 | | Structure of apo outward-open LAT1-CD98h in nanodisc, focused on TMD | | Descriptor: | 4F2 cell-surface antigen heavy chain, Large neutral amino acids transporter small subunit 1 | | Authors: | Lee, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural basis of anticancer drug recognition and amino acid transport by LAT1.
Nat Commun, 16, 2025
|
|
8KDI
 
 | | Structure of apo inward-open LAT1-CD98hc-Fab170 in nanodisc, consensus map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Fab170 heavy chain, ... | | Authors: | Lee, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis of anticancer drug recognition and amino acid transport by LAT1.
Nat Commun, 16, 2025
|
|
8KDF
 
 | | Structure of LAT1-CD98hc in complex with JPH203, focused on TMD | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 4F2 cell-surface antigen heavy chain, CHOLESTEROL, ... | | Authors: | Lee, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Structural basis of anticancer drug recognition and amino acid transport by LAT1.
Nat Commun, 16, 2025
|
|
8KDG
 
 | | Structure of LAT1-CD98hc-Fab170 in complex with BCH, consensus map | | Descriptor: | (1~{S},2~{R},4~{R})-2-azanylbicyclo[2.2.1]heptane-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of anticancer drug recognition and amino acid transport by LAT1.
Nat Commun, 16, 2025
|
|
8KDO
 
 | | Structure of LAT1-CD98hc in complex with melphalan, focused on TMD | | Descriptor: | (2~{S})-2-azanyl-3-[4-[bis(2-chloroethyl)amino]phenyl]propanoic acid, 4F2 cell-surface antigen heavy chain, Large neutral amino acids transporter small subunit 1 | | Authors: | Lee, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural basis of anticancer drug recognition and amino acid transport by LAT1.
Nat Commun, 16, 2025
|
|
1OC2
 
 | |
1QHA
 
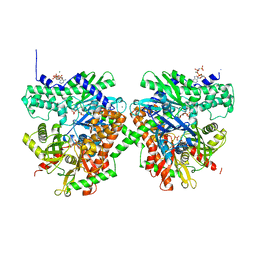 | | HUMAN HEXOKINASE TYPE I COMPLEXED WITH ATP ANALOGUE AMP-PNP | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rosano, C, Sabini, E, Deriu, D, Magnani, M, Bolognesi, M. | | Deposit date: | 1999-05-11 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding of non-catalytic ATP to human hexokinase I highlights the structural components for enzyme-membrane association control.
Structure Fold.Des., 7, 1999
|
|
1GW1
 
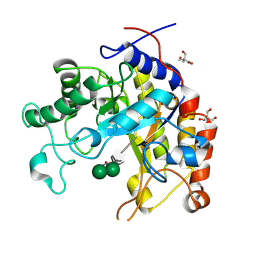 | | Substrate distortion by beta-mannanase from Pseudomonas cellulosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DINITROPHENYLENE, MANNAN ENDO-1,4-BETA-MANNOSIDASE, ... | | Authors: | Ducros, V, Zechel, D.L, Gilbert, H.J, Szabo, L, Withers, S.G, Davies, G.J. | | Deposit date: | 2002-03-01 | | Release date: | 2002-09-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Distortion by a Beta-Mannanase: Snapshots of the Michaelis and Covalent-Intermediate Complexes Suggest a B2,5 Conformation for the Transition State
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
1GVY
 
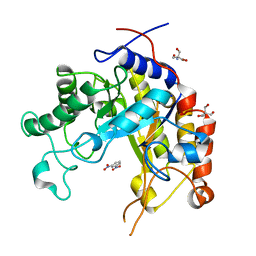 | | Substrate distorsion by beta-mannanase from Pseudomonas cellulosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DINITROPHENYLENE, MANNAN ENDO-1,4-BETA-MANNOSIDASE, ... | | Authors: | Ducros, V, Zechel, D.L, Gilbert, H.J, Szabo, L, Withers, S.G, Davies, G.J. | | Deposit date: | 2002-02-28 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Distortion by a Beta-Mannanase: Snapshots of the Michaelis and Covalent-Intermediate Complexes Suggest a B2,5 Conformation for the Transition State
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
2IQF
 
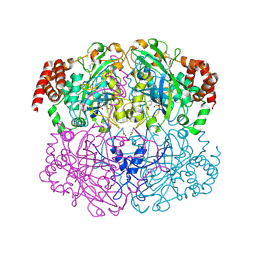 | | Crystal structure of Helicobacter pylori catalase compound I | | Descriptor: | ACETATE ION, Catalase, OXYGEN ATOM, ... | | Authors: | Loewen, P.C, Carpena, X, Fita, I. | | Deposit date: | 2006-10-13 | | Release date: | 2007-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structures and electronic configuration of compound I intermediates of Helicobacter pylori and Penicillium vitale catalases determined by X-ray crystallography and QM/MM density functional theory calculations.
J.Am.Chem.Soc., 129, 2007
|
|
1EQP
 
 | |
1B2H
 
 | |
1B1H
 
 | | OLIGO-PEPTIDE BINDING PROTEIN/TRIPEPTIDE (LYS HPE LYS) COMPLEX | | Descriptor: | PROTEIN (LYS HPE LYS), PROTEIN (OLIGO-PEPTIDE BINDING PROTEIN), URANYL (VI) ION | | Authors: | Davies, T.G, Tame, J.R.H. | | Deposit date: | 1998-11-10 | | Release date: | 1998-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Relating structure to thermodynamics: the crystal structures and binding affinity of eight OppA-peptide complexes.
Protein Sci., 8, 1999
|
|
1B3H
 
 | |
1B5H
 
 | |
1B4H
 
 | |
1B7H
 
 | |
1B6H
 
 | |
1PUY
 
 | | 1.5 A resolution structure of a synthetic DNA hairpin with a stilbenediether linker | | Descriptor: | 5'-D(*GP*TP*TP*TP*TP*GP*(S02)P*CP*AP*AP*AP*AP*C)-3', MAGNESIUM ION | | Authors: | Egli, M, Tereshko, V, Murshudov, G, Sanishvili, R, Liu, X, Lewis, F.D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Face-to-face and edge-to-face pi-pi interactions in a synthetic DNA hairpin with a stilbenediether linker
J.Am.Chem.Soc., 125, 2003
|
|
2OLB
 
 | | OLIGOPEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH TRI-LYSINE | | Descriptor: | ACETATE ION, OLIGO-PEPTIDE BINDING PROTEIN, TRIPEPTIDE LYS-LYS-LYS, ... | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1995-09-10 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands.
Structure, 3, 1995
|
|
