2Z10
 
 | | Crystal structure of putative acetyltransferase | | Descriptor: | Ribosomal-protein-alanine acetyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Genetic Encoding of 3-Iodo-l-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography
Structure, 17, 2009
|
|
2ZXV
 
 | | Crystal structure of putative acetyltransferase from T. thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHA1799 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genetic Encoding of 3-Iodo-l-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography
Structure, 17, 2009
|
|
3AU3
 
 | | Crystal structure of armadillo repeat domain of APC | | Descriptor: | Adenomatous polyposis coli protein | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-28 | | Release date: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the armadillo repeat domain of adenomatous polyposis coli and its complex with the tyrosine-rich domain of sam68
Structure, 19, 2011
|
|
7CNE
 
 | |
3QHE
 
 | | Crystal structure of the complex between the armadillo repeat domain of adenomatous polyposis coli and the tyrosine-rich domain of Sam68 | | Descriptor: | Adenomatous polyposis coli protein, KH domain-containing, RNA-binding, ... | | Authors: | Morishita, E.C.J, Murayama, K, Kato-Murayama, M, Ishizuku-Katsura, Y, Tomabechi, Y, Terada, T, Handa, N, Shirouzu, M, Akiyama, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-25 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the armadillo repeat domain of adenomatous polyposis coli and its complex with the tyrosine-rich domain of sam68
Structure, 19, 2011
|
|
5Y7Z
 
 | | Complex structure of cyclin G-associated kinase with gefitinib | | Descriptor: | Cyclin-G-associated kinase, Gefitinib, NANOBODY, ... | | Authors: | Ohbayashi, N, Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural Basis for the Inhibition of Cyclin G-Associated Kinase by Gefitinib.
ChemistryOpen, 7, 2018
|
|
5Y80
 
 | | Complex structure of cyclin G-associated kinase with gefitinib | | Descriptor: | Cyclin-G-associated kinase, Gefitinib, NANOBODY | | Authors: | Ohbayashi, N, Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Inhibition of Cyclin G-Associated Kinase by Gefitinib.
ChemistryOpen, 7, 2018
|
|
8I5L
 
 | |
2PSM
 
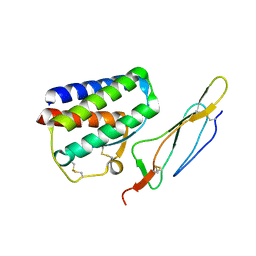 | | Crystal structure of Interleukin 15 in complex with Interleukin 15 receptor alpha | | Descriptor: | BENZAMIDINE, Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Olsen, S.K, Murayama, K, Kishishita, S, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Ota, N, Kanagawa, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Interleukin-15{middle dot}Interleukin-15 Receptor {alpha} Complex: INSIGHTS INTO TRANS AND CIS PRESENTATION
J.Biol.Chem., 282, 2007
|
|
7YM0
 
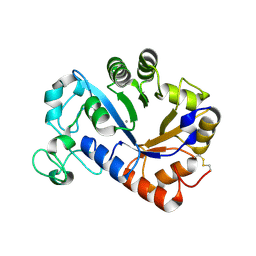 | | Lysoplasmalogen-specific phospholipase D (LyPls-PLD) with Ca2+ | | Descriptor: | CALCIUM ION, Lysoplasmalogenase | | Authors: | Yasutake, Y, Sakasegawa, S, Sugimori, D, Murayama, K. | | Deposit date: | 2022-07-27 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
1UF3
 
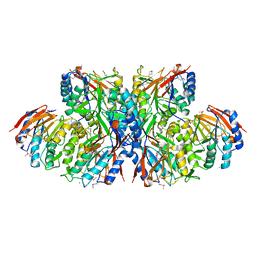 | | Crystal structure of TT1561 of thermus thermophilus HB8 | | Descriptor: | CALCIUM ION, hypothetical protein TT1561 | | Authors: | Kato-Murayama, M, Shirouzu, M, Terada, T, Murayama, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-23 | | Release date: | 2003-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of TT1561 of thermus thermophilus HB8
To be Published
|
|
7JKV
 
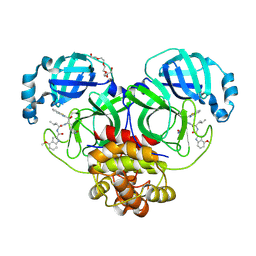 | | Crystal Structure of SARS-CoV-2 main protease in complex with an inhibitor GRL-2420 | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, PENTAETHYLENE GLYCOL | | Authors: | Bulut, H, Hattori, S.I, Das, D, Murayama, K, Mitsuya, H. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication.
Nat Commun, 12, 2021
|
|
9J4G
 
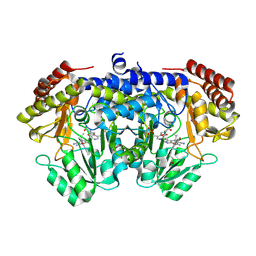 | | Crystal structure of SHMT from E. faecium with (+)-SHIN-2 | | Descriptor: | (+)-SHIN-2, Serine hydroxymethyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Hayashi, H, Murayama, K. | | Deposit date: | 2024-08-09 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SHIN-2 exerts potent activity against VanA-type vancomycin-resistant Enterococcus faecium in vitro by stabilizing the active site loop of serine hydroxymethyltransferase.
Arch.Biochem.Biophys., 761, 2024
|
|
1WEK
 
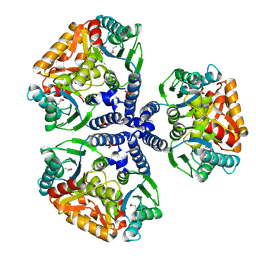 | | Crystal structure of the conserved hypothetical protein TT1465 from Thermus thermophilus HB8 | | Descriptor: | PHOSPHATE ION, hypothetical protein TT1465 | | Authors: | Kukimoto-Niino, M, Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of possible lysine decarboxylases from Thermus thermophilus HB8
Protein Sci., 13, 2004
|
|
1J27
 
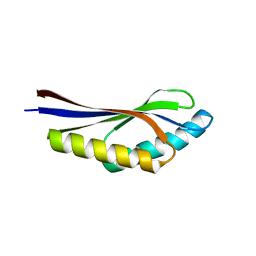 | | Crystal structure of a hypothetical protein, TT1725, from Thermus thermophilus HB8 at 1.7A resolution | | Descriptor: | hypothetical protein TT1725 | | Authors: | Seto, A, Shirouzu, M, Terada, T, Murayama, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-26 | | Release date: | 2003-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a hypothetical protein, TT1725, from Thermus thermophilus HB8 at 1.7 A resolution
Proteins, 53, 2003
|
|
2CWK
 
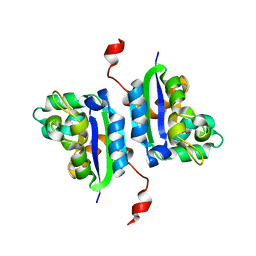 | |
2DXF
 
 | | Crystal structure of nucleoside diphosphate kinase in complex with GTP analog | | Descriptor: | CHLORIDE ION, Nucleoside diphosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-25 | | Release date: | 2007-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of nucleoside diphosphate kinase in complex with GTP analog
To be Published
|
|
2DY9
 
 | | Crystal structure of nucleoside diphosphate kinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-08 | | Release date: | 2007-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of nucleoside diphosphate kinase in complex with ADP
To be Published
|
|
2DXA
 
 | | Crystal structure of trans editing enzyme ProX from E.coli | | Descriptor: | CHLORIDE ION, Protein ybaK | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-25 | | Release date: | 2007-02-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of trans editing enzyme ProX from E.coli
To be Published
|
|
2DYC
 
 | | Crystal structure of the N-terminal domain of mouse galectin-4 | | Descriptor: | Galectin-4 | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-08 | | Release date: | 2007-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the N-terminal domain of mouse galectin-4
To be Published
|
|
2CYE
 
 | | Crystal structure of Thioesterase complexed with coenzyme A and Zn from Thermus thermophilus HB8 | | Descriptor: | COENZYME A, Putative thioesterase, ZINC ION | | Authors: | Hosaka, T, Kato-Murayama, M, Murayama, K, Kishishita, S, Handa, N, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2006-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the putative thioesterase protein TTHA1846 from Thermus thermophilus HB8 complexed with coenzyme A and a zinc ion.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2DXD
 
 | | Crystal structure of nucleoside diphosphate kinase in complex with ATP analog | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Nucleoside diphosphate kinase | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-25 | | Release date: | 2007-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of nucleoside diphosphate kinase in complex with ATP analog
To be Published
|
|
2DYA
 
 | | Crystal structure of complex between Adenine nucleotide and nucleoside diphosphate kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-08 | | Release date: | 2007-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of complex between Adenine nucleotide and nucleoside diphosphate kinase
To be Published
|
|
2DXE
 
 | | Crystal structure of nucleoside diphosphate kinase in complex with GDP | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-25 | | Release date: | 2007-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of nucleoside diphosphate kinase in complex with GDP
To be Published
|
|
1WEH
 
 | | Crystal structure of the conserved hypothetical protein TT1887 from Thermus thermophilus HB8 | | Descriptor: | Conserved hypothetical protein TT1887 | | Authors: | Kukimoto-Niino, M, Murayama, K, Idaka, M, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of possible lysine decarboxylases from Thermus thermophilus HB8
PROTEIN SCI., 13, 2004
|
|
