5YZ7
 
 | | Crystal structure of OsD14 in complex with D-ring-opened 7'-carba-4BD | | Descriptor: | (2Z,4S)-5-(4-bromophenyl)-4-hydroxy-2-methylpent-2-enoic acid, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Jiang, K, Xu, Y, Miyakawa, T, Asami, T, Tanokura, M. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Rationally Designed Strigolactone Analogs as Antagonists of the D14 Receptor.
Plant Cell Physiol., 59, 2018
|
|
5ZHR
 
 | | Crystal structure of OsD14 in complex with covalently bound KK094 | | Descriptor: | (2,3-dihydro-1H-indol-1-yl)(1H-1,2,3-triazol-1-yl)methanone, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
5ZHT
 
 | | Crystal structure of OsD14 in complex with covalently bound KK073 | | Descriptor: | (1H-1,2,3-triazol-1-yl){4-[4-(trifluoromethyl)phenyl]piperazin-1-yl}methanone, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
6AGZ
 
 | | Crystal structure of Old Yellow Enzyme from Pichia sp. AKU4542 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old Yellow Enzyme | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2018-08-15 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of different substrate preferences of two old yellow enzymes from yeasts in the asymmetric reduction of enone compounds.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
6JH5
 
 | | Structure of Marine bacterial laminarinase | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Ru, L, Tanokura, M, Long, L. | | Deposit date: | 2019-02-17 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6JIA
 
 | | Marine bacterial laminarinase mutant E135A complex with laminaritetraose | | Descriptor: | CALCIUM ION, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, laminarinase | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6M6P
 
 | | Structure of Marine bacterial laminarinase mutant E135A in complex with 1,3-beta-cellotriosyl-glucose | | Descriptor: | CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, laminarinase | | Authors: | Yang, J, Xu, Y, Tanokura, M, Long, L, Miyakawa, T. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6JHJ
 
 | | Structure of Marine bacterial laminarinase mutant-E135A | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
7VN7
 
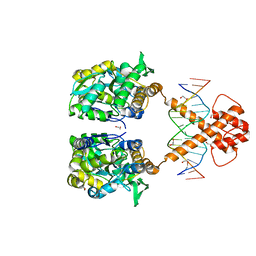 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning GACACGTGTC | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*GP*AP*CP*AP*CP*GP*TP*GP*TP*CP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN8
 
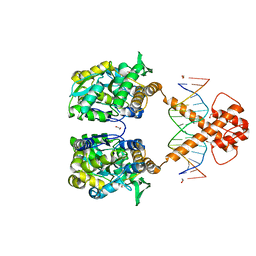 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning GTCACGTGAC | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*GP*TP*CP*AP*CP*GP*TP*GP*AP*CP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN3
 
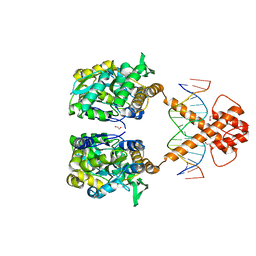 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning CACACGTGTG | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN6
 
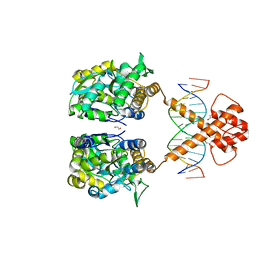 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning CGCACGTGCG | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*GP*CP*AP*CP*GP*TP*GP*CP*GP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN4
 
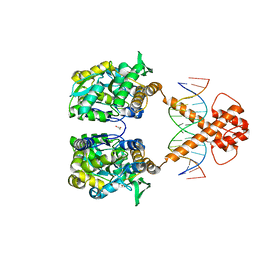 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning TCCACGTGGA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN5
 
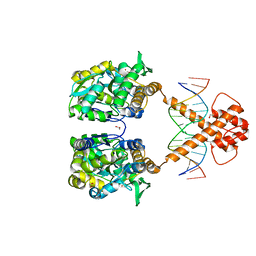 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning TTCACGTGAA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*TP*CP*AP*CP*GP*TP*GP*AP*AP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN2
 
 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning ATCACGTGAT | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*AP*TP*CP*AP*CP*GP*TP*GP*AP*TP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
3W20
 
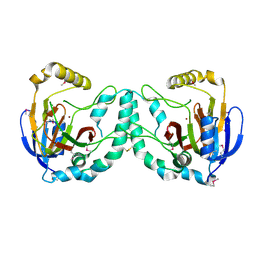 | | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Qin, H.M, Miyakawa, T, Jia, M.Z, Nakamura, A, Ohtsuka, J, Xue, Y.L, Kawashima, T, Kasahara, T, Hibi, M, Ogawa, J, Tanokura, M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD
Plos One, 8, 2013
|
|
3W21
 
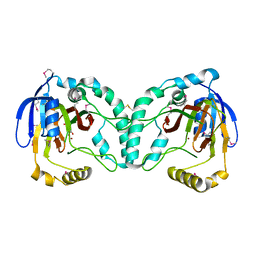 | | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase in complex with alpha-KG from Burkholderia ambifaria AMMD | | Descriptor: | 2-OXOGLUTARIC ACID, Putative uncharacterized protein, ZINC ION | | Authors: | Qin, H.M, Miyakawa, T, Jia, M.Z, Nakamura, A, Ohtsuka, J, Xue, Y.L, Kawashima, T, Kasahara, T, Hibi, M, Ogawa, J, Tanokura, M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD
Plos One, 8, 2013
|
|
3VXK
 
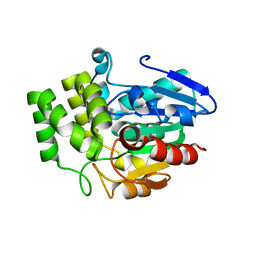 | | Crystal structure of OsD14 | | Descriptor: | Dwarf 88 esterase | | Authors: | Xue, Y.-L, Miyakawa, T, Hou, F, Qin, H.-M, Tanokura, M. | | Deposit date: | 2012-09-18 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanism of strigolactone perception by DWARF14
Nat Commun, 4, 2013
|
|
3WIO
 
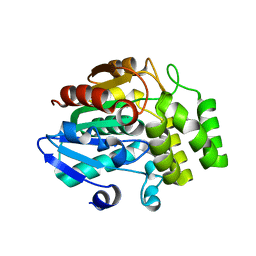 | | Crystal structure of OSD14 in complex with hydroxy D-ring | | Descriptor: | (5R)-5-hydroxy-3-methylfuran-2(5H)-one, Probable strigolactone esterase D14 | | Authors: | Xue, Y.-L, Miyakawa, T, Hou, F, Qin, H.-M, Tanokura, M. | | Deposit date: | 2013-09-22 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of strigolactone perception by DWARF14
Nat Commun, 4, 2013
|
|
3VXG
 
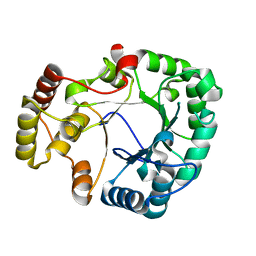 | | Crystal structure of conjugated polyketone reductase C2 from Candida Parapsilosis | | Descriptor: | Conjugated polyketone reductase C2 | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WAZ
 
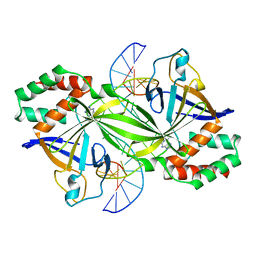 | | Crystal structure of a restriction enzyme PabI in complex with DNA | | Descriptor: | ADENINE, DNA (5'-D(*GP*CP*AP*TP*AP*GP*CP*TP*GP*TP*(ORP)P*CP*AP*GP*CP*TP*AP*TP*GP*C)-3'), Putative uncharacterized protein | | Authors: | Miyazono, K, Miyakawa, T, Ito, T, Tanokura, M. | | Deposit date: | 2013-05-10 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A sequence-specific DNA glycosylase mediates restriction-modification in Pyrococcus abyssi.
Nat Commun, 5, 2014
|
|
3WWH
 
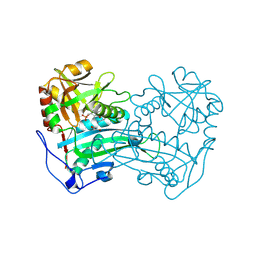 | | Crystal structure of the first R-stereoselective -transaminase identified from Arthrobacter sp. KNK168 (FERM-BP-5228) | | Descriptor: | (R)-amine transaminase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3WWI
 
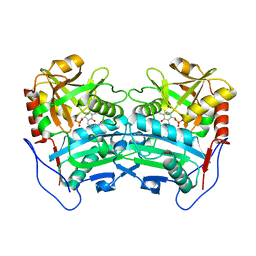 | | Crystal structure of the G136F mutant of the first R-stereoselective -transaminase identified from Arthrobacter sp. KNK168 (FERM-BP-5228) | | Descriptor: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Miyakawa, T, Zhi, Y, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3WQB
 
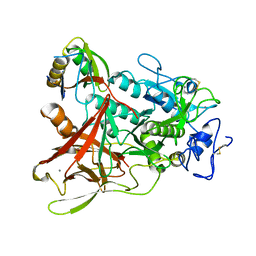 | | Crystal structure of aeromonas sobria serine protease (ASP) and the chaperone (ORF2) complex | | Descriptor: | CALCIUM ION, Extracellular serine protease, Open reading frame 2 | | Authors: | Kobayashi, H, Yoshida, T, Miyakawa, T, Kato, R, Tashiro, M, Yamanaka, H, Tanokura, M, Tsuge, H. | | Deposit date: | 2014-01-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Basis for Action of the External Chaperone for a Propeptide-deficient Serine Protease from Aeromonas sobria.
J.Biol.Chem., 290, 2015
|
|
3WWJ
 
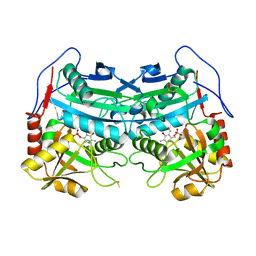 | | Crystal structure of an engineered sitagliptin-producing transaminase, ATA-117-Rd11 | | Descriptor: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Hou, F, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
