6F3X
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide in frozen DDM/CHS detergent micelle solution determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3Y
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide bound to human Bradykinin 1 Receptor (B1R) determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
1KHE
 
 | | PEPCK complex with nonhydrolyzable GTP analog, MAD data | | Descriptor: | MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Phosphoenolpyruvate Carboxykinase, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
1KHB
 
 | | PEPCK complex with nonhydrolyzable GTP analog, native data | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
1KHG
 
 | | PEPCK | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
1KHF
 
 | | PEPCK complex with PEP | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
6HFB
 
 | | Outward-facing conformation of a multidrug resistance MATE family transporter of the MOP superfamily. | | Descriptor: | CESIUM ION, Uncharacterized protein | | Authors: | Nonaka, T, Zakrzewska, S, Safarian, S, Michel, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Inward-facing conformation of a multidrug resistance MATE family transporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1PCR
 
 | | STRUCTURE OF THE PHOTOSYNTHETIC REACTION CENTRE FROM RHODOBACTER SPHAEROIDES AT 2.65 ANGSTROMS RESOLUTION: COFACTORS AND PROTEIN-COFACTOR INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Ermler, U, Fritzsch, G, Michel, H. | | Deposit date: | 1994-11-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.65 A resolution: cofactors and protein-cofactor interactions.
Structure, 2, 1994
|
|
1QLE
 
 | |
6FHZ
 
 | |
6GWH
 
 | |
1EZV
 
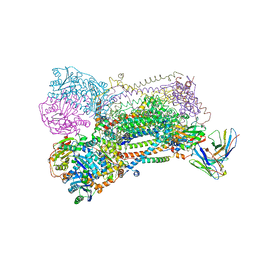 | | STRUCTURE OF THE YEAST CYTOCHROME BC1 COMPLEX CO-CRYSTALLIZED WITH AN ANTIBODY FV-FRAGMENT | | Descriptor: | 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CYTOCHROME B, CYTOCHROME C1, ... | | Authors: | Hunte, C, Koepke, J, Lange, C, Rossmanith, T, Michel, H. | | Deposit date: | 2000-05-12 | | Release date: | 2001-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment.
Structure Fold.Des., 8, 2000
|
|
2JBL
 
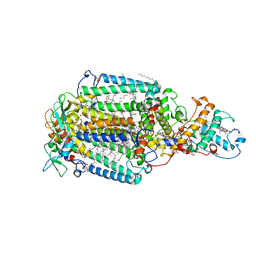 | | PHOTOSYNTHETIC REACTION CENTER FROM BLASTOCHLORIS VIRIDIS | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2006-12-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Comparison of Stigmatellin Conformations, Free and Bound to the Photosynthetic Reaction Center and the Cytochrome Bc(1) Complex.
J.Mol.Biol., 368, 2007
|
|
6KLV
 
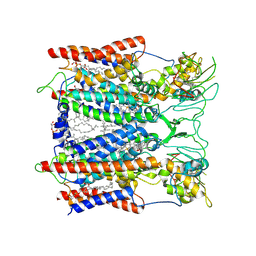 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ANTIMYCIN, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7DEG
 
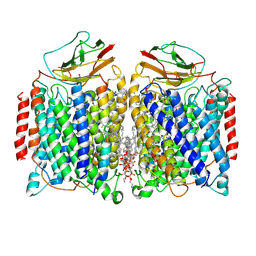 | | Cryo-EM structure of a heme-copper terminal oxidase dimer provides insights into its catalytic mechanism | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guoliang, Z, Shuangbo, Z. | | Deposit date: | 2020-11-04 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Unusual Homodimer of a Heme-Copper Terminal Oxidase Allows Itself to Utilize Two Electron Donors.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3EHB
 
 | | A D-Pathway Mutation Decouples the Paracoccus Denitrificans Cytochrome c Oxidase by Altering the side chain orientation of a distant, conserved Glutamate | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cytochrome c oxidase subunit 1-beta, ... | | Authors: | Koepke, J, Mueller, H, Peng, G. | | Deposit date: | 2008-09-12 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A d-pathway mutation decouples the paracoccusdenitrificans cytochrome C oxidase by altering the side-chain orientation of a distant conserved glutamate
J.Mol.Biol., 384, 2008
|
|
1M51
 
 | | PEPCK complex with a GTP-competitive inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Foley, L.H, Wang, P, Dunten, P, Wertheimer, S.J. | | Deposit date: | 2002-07-06 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray Structures of two xanthine inhibitors bound to PEPCK and N-3 modifications of substituted 1,8-Dibenzylxanthines
Bioorg.Med.Chem.Lett., 13, 2003
|
|
5OVQ
 
 | |
4RCR
 
 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
7QKS
 
 | |
7QKR
 
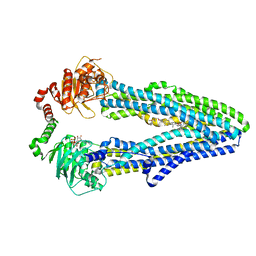 | | Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris with Verapamil at 3.2 A resolution | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, Dexverapamil, MAGNESIUM ION, ... | | Authors: | Schleker, E.S.M, Reinhart, C. | | Deposit date: | 2021-12-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional investigation of ABC transporter STE6-2p from Pichia pastoris reveals unexpected interaction with sterol molecules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3HB3
 
 | | High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase | | Descriptor: | ANTIBODY FV FRAGMENT, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Koepke, J, Angerer, H, Peng, G. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: New insights into the active site and the proton transfer pathways
Biochim.Biophys.Acta, 1787, 2009
|
|
7Z6Q
 
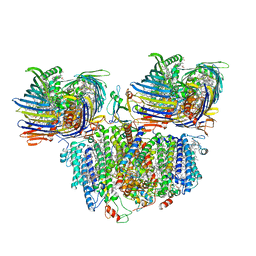 | |
2BS2
 
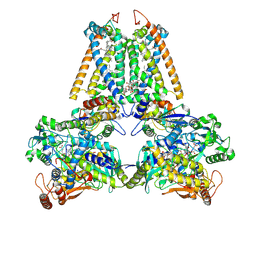 | | QUINOL:FUMARATE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2005-05-14 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Evidence for Transmembrane Proton Transfer in a Dihaem-Containing Membrane Protein Complex.
Embo J., 25, 2006
|
|
2BS4
 
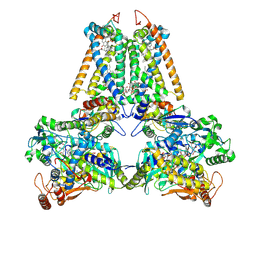 | | GLU C180 -> ILE VARIANT QUINOL:FUMARATE REDUCTASE FROMWOLINELLA SUCCINOGENES | | Descriptor: | 2,3-DIMETHYL-1,4-NAPHTHOQUINONE, CITRIC ACID, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2005-05-14 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Experimental Support for the E-Pathway Hypothesis of Coupled Transmembrane Electron and Proton Transfer in Dihemic Quinol:Fumarate Reductase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
