5B0B
 
 | | Polyketide cyclase OAC from Cannabis sativa, I7F mutant | | Descriptor: | ACETATE ION, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
1JWI
 
 | | Crystal Structure of Bitiscetin, a von Willeband Factor-dependent Platelet Aggregation Inducer. | | Descriptor: | bitiscetin, platelet aggregation inducer | | Authors: | Hirotsu, S, Mizuno, H, Fukuda, K, Qi, M.C, Matsui, T, Hamako, J, Morita, T, Titani, K. | | Deposit date: | 2001-09-04 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of bitiscetin, a von Willebrand factor-dependent platelet aggregation inducer.
Biochemistry, 40, 2001
|
|
7EQE
 
 | | Crystal Structure of a transcription factor | | Descriptor: | TetR/AcrR family transcriptional regulator | | Authors: | Uehara, S, Tsugita, A, Matsui, T, Yokoyama, T, Ostash, I, Ostash, B, Tanaka, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK.
Febs J., 289, 2022
|
|
7EQF
 
 | | Crystal Structure of a Transcription Factor in complex with Ligand | | Descriptor: | (6~{R})-3-methyl-8-[(2~{S},4~{R},5~{S},6~{R})-6-methyl-5-[(2~{S},4~{R},5~{R},6~{R})-6-methyl-4-[(2~{S},5~{S},6~{S})-6-methyl-5-[(2~{S},4~{R},5~{S},6~{R})-6-methyl-5-[(2~{S},4~{S},5~{S},6~{R})-6-methyl-4-[(2~{S},5~{S},6~{S})-6-methyl-5-oxidanyl-oxan-2-yl]oxy-5-oxidanyl-oxan-2-yl]oxy-4-oxidanyl-oxan-2-yl]oxy-oxan-2-yl]oxy-5-oxidanyl-oxan-2-yl]oxy-4-oxidanyl-oxan-2-yl]oxy-1,6,11-tris(oxidanyl)-5,6-dihydrobenzo[a]anthracene-7,12-dione, TetR/AcrR family transcriptional regulator | | Authors: | Uehara, S, Tsugita, A, Matsui, T, Yokoyama, T, Ostash, I, Ostash, B, Tanaka, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK.
Febs J., 289, 2022
|
|
3WT0
 
 | |
3WD7
 
 | | Type III polyketide synthase | | Descriptor: | COENZYME A, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Shimokawa, Y, Matsui, T, Kato, R, Sugio, S, Morita, H, Abe, I. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
3WD8
 
 | | TypeIII polyketide synthases | | Descriptor: | GLYCEROL, Type III polyketide synthase quinolone synthase | | Authors: | Mori, T, Shimokawa, Y, Matsui, T, Morita, H, Abe, I. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
3WXZ
 
 | | The structure of the I375F mutant of CsyB | | Descriptor: | Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
3WY0
 
 | | The I375W mutant of CsyB complexed with CoA-SH | | Descriptor: | COENZYME A, Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
3WXY
 
 | | Crystal structure of CsyB complexed with CoA-SH | | Descriptor: | COENZYME A, Putative uncharacterized protein csyB | | Authors: | Mori, T, Yang, D, Matsui, T, Morita, H, Fujii, I, Abe, I. | | Deposit date: | 2014-08-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Structural basis for the formation of acylalkylpyrones from two beta-ketoacyl units by the fungal type III polyketide synthase CsyB.
J.Biol.Chem., 290, 2015
|
|
1OFK
 
 | |
1OFJ
 
 | |
7XVJ
 
 | | Crystal structure of CdpNPT in complex with harmol | | Descriptor: | 1-methyl-9~{H}-pyrido[3,4-b]indol-7-ol, Cyclic dipeptide N-prenyltransferase, PHOSPHATE ION | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-05-24 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzymatic formation of a prenyl beta-carboline by a fungal indole prenyltransferase.
J Nat Med, 76, 2022
|
|
4G56
 
 | | Crystal Structure of full length PRMT5/MEP50 complexes from Xenopus laevis | | Descriptor: | Hsl7 protein, MGC81050 protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ho, M, Wilczek, C, Bonanno, J, Shechter, D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity
Plos One, 8, 2013
|
|
4IFQ
 
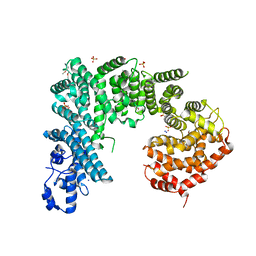 | | Crystal structure of Saccharomyces cerevisiae NUP192, residues 2 to 960 [ScNup192(2-960)] | | Descriptor: | IODIDE ION, Nucleoporin NUP192, SULFATE ION | | Authors: | Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2012-12-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex.
Structure, 21, 2013
|
|
4X41
 
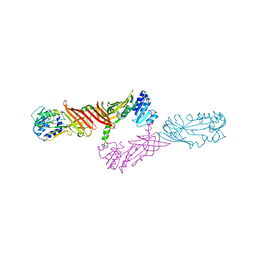 | | Crystal Structure of Protein Arginine Methyltransferase PRMT8 | | Descriptor: | Protein arginine N-methyltransferase 8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lee, W.C, Ho, M.C. | | Deposit date: | 2014-12-02 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Protein Arginine Methyltransferase 8: Tetrameric Structure and Protein Substrate Specificity
Biochemistry, 54, 2015
|
|
4XKH
 
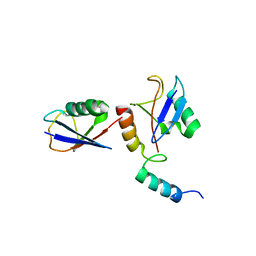 | | CRYSTAL STRUCTURE OF THE AIRAPL TANDEM UIMS IN COMPLEX WITH A LYS48-LINKED TRI-UBIQUITIN | | Descriptor: | AN1-type zinc finger protein 2B, Polyubiquitin-C | | Authors: | Rahighi, S, Kawasaki, M, Stanhill, A, Wakatsuki, S. | | Deposit date: | 2015-01-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective Binding of AIRAPL Tandem UIMs to Lys48-Linked Tri-Ubiquitin Chains.
Structure, 24, 2016
|
|
1O16
 
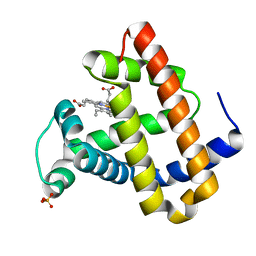 | | RECOMBINANT SPERM WHALE MYOGLOBIN H64D/V68S/D122N MUTANT (MET) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2002-10-25 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular engineering of myoglobin: influence of residue 68 on the rate and the
enantioselectivity of oxidation reactions catalyzed by H64D/V68X myoglobin
Biochemistry, 42, 2003
|
|
7SEH
 
 | | Glucose-6-phosphate 1-dehydrogenase (K403QdLtL) | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mathews, I.I, Garcia, A.A, Wakatsuki, S, Mochly-Rosen, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Stabilization of glucose-6-phosphate dehydrogenase oligomers enhances catalytic activity and stability of clinical variants.
J.Biol.Chem., 298, 2022
|
|
7SEI
 
 | | Glucose-6-phosphate 1-dehydrogenase (K403Q) | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mathews, I.I, Garcia, A.A, Wakatsuki, S, Mochly-Rosen, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Stabilization of glucose-6-phosphate dehydrogenase oligomers enhances catalytic activity and stability of clinical variants.
J.Biol.Chem., 298, 2022
|
|
3NF5
 
 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
6OQ7
 
 | | Structure of the GTD domain of Clostridium difficile toxin B in complex with VHH E3 | | Descriptor: | E3, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OQ8
 
 | |
6DKK
 
 | | Structure of BoNT | | Descriptor: | Botulinum neurotoxin type A, PHOSPHATE ION | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2018-05-29 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A viral-fusion-peptide-like molecular switch drives membrane insertion of botulinum neurotoxin A1.
Nat Commun, 9, 2018
|
|
6OQ6
 
 | |
